Immobilized pH gradient isoelectric focusing as a first-dimension separation in shotgun proteomics
- PMID: 16461941
- PMCID: PMC2291737
Immobilized pH gradient isoelectric focusing as a first-dimension separation in shotgun proteomics
Abstract
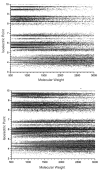
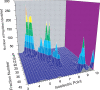
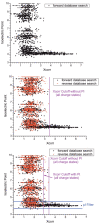
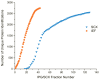
Shotgun proteomics, where a tryptic digest of a complex proteome sample is directly analyzed by either single dimensional or multidimensional liquid chromatography tandem mass spectrometry, has gained acceptance in the proteomics community at large and is widely used in core facilities. Here we review the development in our laboratory of an alternative first-dimension separation technique for shotgun proteomics, immobilized pH gradient isoelectric focusing (IPG-IEF). The key advantages of the technology over other multidimensional separation formats (simplicity, high resolution, and high sensitivity) are discussed. The concept of using peptide pI to filter large shotgun proteomics datasets generated by the IPG-IEF technique to minimize false positives and negatives is also introduced. Finally, an account of the comparison of the technique with the established gold standard for multidimensional separation of peptides, strong cation exchange chromatography, is presented, along with the prospects for the use of peptide pI along with accurate mass measurement for the identification of peptides.
Figures


References
-
- Rejtar T, Hu P, Juhasz P, et al. Off-line coupling of high-resolution capillary electrophoresis to MALDI-TOF and TOF/TOF MS. J Proteome Res 2002;1:171–179. - PubMed
-
- Washburn MP, Wolters D, Yates JR, 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 2001; 19:242–247. - PubMed
-
- Peng J, Elias JE, Thoreen CC, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J Proteome Res 2003; 2:43–50. - PubMed
-
- Egen NB, Bliss M, Mayersohn M, et al. Isolation of monoclonal antibodies to phencyclidine from ascites fluid by preparative isoelectric focusing in the Rotofor. Anal Biochem 1988;172:488–494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
