Optimization of in vitro transcription and full-length cDNA synthesis using the T4 bacteriophage gene 32 protein
- PMID: 16461948
- PMCID: PMC2291727
Optimization of in vitro transcription and full-length cDNA synthesis using the T4 bacteriophage gene 32 protein
Abstract
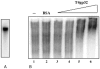
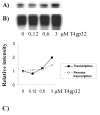
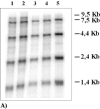
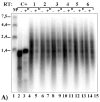
We evaluated the effect of the T4 bacteriophage gene 32 protein (T4gp32) on in vitro transcription and reverse transcription. T4gp32 doubled the yield of in vitro transcripts obtained with T7 RNA polymerase and increased the yield of cDNA synthesis when used in combination with an RNaseH-deficient Moloney murine leukemia virus [Au: ok] reverse transcriptase. The positive effect could be correlated with the RNA chaperone activity of T4gp32. T4gp32 stimulated the synthesis of long cDNAs, particularly species longer than 7 kb. By comparison, thermal activation of reverse transcriptase with trehalose only boosted the production of shorter cDNAs. For the construction of an Arabidopsis thaliana cDNA library, where the average cDNA size is 1.2 kbp, both the presence of T4gp32 under standard reaction conditions as well as thermal activation resulted in similarly high percentages of full-length cDNA. However, the inclusion of T4gp32 in a standard reverse transcription reaction resulted in the highest cDNA yield. We conclude that the addition of T4gp32 in standard reverse transcription reactions can increase the quality and yield of full-length cDNA libraries.
Figures


Similar articles
-
A highly efficient method for long-chain cDNA synthesis using trehalose and betaine.Anal Biochem. 2002 Feb 15;301(2):168-74. doi: 10.1006/abio.2001.5474. Anal Biochem. 2002. PMID: 11814287
-
Construction of infectious clones for RNA viruses: TMV.Methods Mol Biol. 2008;451:477-90. doi: 10.1007/978-1-59745-102-4_32. Methods Mol Biol. 2008. PMID: 18370275
-
Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5'-RACE and primer extension.Biotechniques. 2001 Mar;30(3):574-80, 582. doi: 10.2144/01303rr02. Biotechniques. 2001. PMID: 11252793
-
Synthesis in high yield of complementary DNA of retroviral RNA.Prep Biochem. 1980;10(4):483-93. doi: 10.1080/00327488008061745. Prep Biochem. 1980. PMID: 6158040
-
Moloney murine leukemia reverse transcriptase suspect in the production of multiple misincorporations during hprt cDNA synthesis.Mutat Res. 1997 Mar 4;374(1):145-8. doi: 10.1016/s0027-5107(96)00251-5. Mutat Res. 1997. PMID: 9067424
Cited by
-
Isothermal Amplification of Long, Discrete DNA Fragments Facilitated by Single-Stranded Binding Protein.Sci Rep. 2017 Aug 17;7(1):8497. doi: 10.1038/s41598-017-09063-x. Sci Rep. 2017. PMID: 28819114 Free PMC article.
-
Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription.J Virol. 2007 Sep;81(18):10037-46. doi: 10.1128/JVI.00519-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626089 Free PMC article.
-
Single-molecule stretching studies of RNA chaperones.RNA Biol. 2010 Nov-Dec;7(6):712-23. doi: 10.4161/rna.7.6.13776. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045548 Free PMC article. Review.
-
A comparison of RNA amplification techniques at sub-nanogram input concentration.BMC Genomics. 2009 Jul 20;10:326. doi: 10.1186/1471-2164-10-326. BMC Genomics. 2009. PMID: 19619282 Free PMC article.
-
Protocol for nearly full-length sequencing of HIV-1 RNA from plasma.PLoS One. 2008 Jan 9;3(1):e1420. doi: 10.1371/journal.pone.0001420. PLoS One. 2008. PMID: 18183300 Free PMC article.
References
-
- Sugahara Y, Carninci P, Itoh M, et al. Comparative evaluation of 5′-end-sequence quality of clones in CAP trapper and other full-length-cDNA libraries. Gene 2001; 263:93–102. - PubMed
-
- Sambrook J, Russel DW. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001.
-
- Carninci P, Shiraki T, Mizuno Y, Muramatsu M, Hayashizaki Y. Extra-long first-strand cDNA synthesis. BioTechniques 2002;32:984–985. - PubMed
-
- Demeke T, Morris F. Molecular characterization of wheat polyphenol oxidase (PPO). Theor Appl Genet 2002;104:813–818. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
