Direct reversal of DNA alkylation damage
- PMID: 16464003
- PMCID: PMC2432087
- DOI: 10.1021/cr0404702
Direct reversal of DNA alkylation damage
Figures

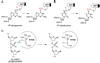
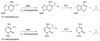
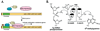
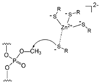


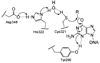


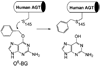
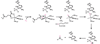



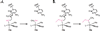
References
-
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995.
-
- Lindahl T. Nature. Vol. 362. 1993. p. 709. - PubMed
-
- Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE. DNA Repair. 2004;3:1389. - PubMed
-
- Vaughan P, Lindahl T, Sedgwick B. Mutat. Res. 1993;293:249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

