A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol
- PMID: 16465619
- PMCID: PMC1380285
- DOI: 10.1086/500615
A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol
Abstract
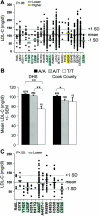
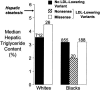
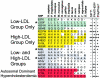
Selected missense mutations in the proprotein convertase subtilisin/kexin type 9 serine protease gene (PCSK9) cause autosomal dominant hypercholesterolemia, whereas nonsense mutations in the same gene are associated with low plasma levels of low-density lipoprotein cholesterol (LDL-C). Here, DNA sequencing and chip-based oligonucleotide hybridization were used to determine whether other sequence variations in PCSK9 contribute to differences in LDL-C levels. The coding regions of PCSK9 were sequenced in the blacks and whites from the Dallas Heart Study (n=3,543) who had the lowest (<5th percentile) and highest (>95th percentile) plasma levels of LDL-C. Of the 17 missense variants identified, 3 (R46L, L253F, and A443T) were significantly and reproducibly associated with lower plasma levels of LDL-C (reductions ranging from 3.5% to 30%). None of the low-LDL-C variants were associated with increased hepatic triglyceride content, as measured by proton magnetic resonance spectroscopy. This finding is most consistent with the reduction in LDL-C being caused primarily by accelerating LDL clearance, rather than by reduced lipoprotein production. Association studies with 93 noncoding single-nucleotide polymorphisms (SNPs) at the PCSK9 locus identified 3 SNPs associated with modest differences in plasma LDL-C levels. Thus, a spectrum of sequence variations ranging in frequency (from 0.2% to 34%) and magnitude of effect (from a 3% increase to a 49% decrease) contribute to interindividual differences in LDL-C levels. These findings reveal that PCSK9 activity is a major determinant of plasma levels of LDL-C in humans and make it an attractive therapeutic target for LDL-C lowering.
Figures



References
Web Resources
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for LDLR, autosomal dominant hypercholesterolemia, and autosomal recessive hypercholesterolemia)
-
- Sorting Intolerant From Tolerant (SIFT), http://blocks.fhcrc.org/sift/SIFT.html
-
- The R Project for Statistical Computing, http://www.r-project.org/
References
-
- Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the LDLR and LDL-cholesterol. J Biol Chem 279:48865–48875 10.1074/jbc.M409699200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

