Rac1 and Cdc42 have different roles in Candida albicans development
- PMID: 16467473
- PMCID: PMC1405900
- DOI: 10.1128/EC.5.2.321-329.2006
Rac1 and Cdc42 have different roles in Candida albicans development
Abstract
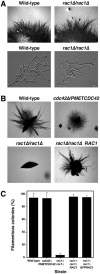
We investigated the role of the highly conserved G protein Rac1 in the opportunistic pathogen Candida albicans. We identified and disrupted RAC1 and show here that, in contrast to CDC42, it is not necessary for viability or serum-induced hyphal growth but is essential for filamentous growth when cells are embedded in a matrix. Rac1 is localized to the plasma membrane, yet its distribution is more homogenous than that of Cdc42, with no enrichment at the tips of either buds or hyphae. In addition, fluorescence recovery after photobleaching results indicate that Rac1 and Cdc42 have different dynamics at the membrane. Furthermore, overexpression of Rac1 does not complement Cdc42 function, and conversely, overexpression of Cdc42 does not complement Rac1 function. Thus, Rac1 and Cdc42, although highly similar to one another, have different roles in C. albicans development.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2003. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 116:1249-1260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

