Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer
- PMID: 16467855
- PMCID: PMC1422165
- DOI: 10.1038/sj.emboj.7600969
Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer
Abstract
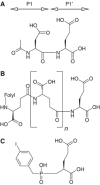
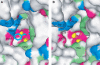
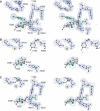
Membrane-bound glutamate carboxypeptidase II (GCPII) is a zinc metalloenzyme that catalyzes the hydrolysis of the neurotransmitter N-acetyl-L-aspartyl-L-glutamate (NAAG) to N-acetyl-L-aspartate and L-glutamate (which is itself a neurotransmitter). Potent and selective GCPII inhibitors have been shown to decrease brain glutamate and provide neuroprotection in preclinical models of stroke, amyotrophic lateral sclerosis, and neuropathic pain. Here, we report crystal structures of the extracellular part of GCPII in complex with both potent and weak inhibitors and with glutamate, the product of the enzyme's hydrolysis reaction, at 2.0, 2.4, and 2.2 A resolution, respectively. GCPII folds into three domains: protease-like, apical, and C-terminal. All three participate in substrate binding, with two of them directly involved in C-terminal glutamate recognition. One of the carbohydrate moieties of the enzyme is essential for homodimer formation of GCPII. The three-dimensional structures presented here reveal an induced-fit substrate-binding mode of this key enzyme and provide essential information for the design of GCPII inhibitors useful in the treatment of neuronal diseases and prostate cancer.
Figures


References
-
- Anilkumar G, Rajasekaran SA, Wang S, Hankinson O, Bander NH, Rajasekaran AK (2003) Prostate-specific membrane antigen association with filamin A modulates its internalization and NAALADase activity. Cancer Res 63: 2645–2648 - PubMed
-
- Barinka C, Rinnova M, Sacha P, Rojas C, Majer P, Slusher BS, Konvalinka J (2002) Substrate specificity, inhibition and enzymological analysis of recombinant human glutamate carboxypeptidase II. J Neurochem 80: 477–487 - PubMed
-
- Bennett MJ, Lebron JA, Bjorkman PJ (2000) Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403: 46–53 - PubMed
-
- Benveniste H, Drejer J, Schousboe A, Diemer NH (1984) Elevation of the extracellular concentrations of glutamate and aspartate in rat hippocampus during transient cerebral ischemia monitored by intracerebral microdialysis. J Neurochem 43: 1369–1374 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

