Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity
- PMID: 16469845
- PMCID: PMC1413832
- DOI: 10.1073/pnas.0511138103
Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity
Abstract
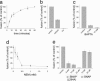
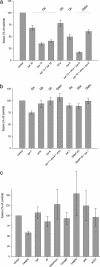
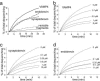
Membrane fusion in the secretory pathway is mediated by soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins. Different fusion steps are thought to be effected by independent sets of SNAREs, but it is unclear whether specificity is determined by an intrinsic specificity of SNARE pairing or by upstream factors. Using a newly developed microscopy-based assay, we have investigated the SNARE specificity of homotypic early endosomal fusion. We show that early endosomes contain multiple sets of SNAREs, including, in addition to the putative early endosomal SNAREs, those involved in exocytosis and in fusion of late endosomes. We demonstrate that fusion is largely mediated by a complex formed by syntaxin 13, syntaxin 6, vti1a, and VAMP4, whereas the exocytic and late endosomal SNAREs play little or no role in the reaction. In contrast, proteoliposomes reconstituted with early endosomal SNAREs promiscuously fuse with liposomes containing exocytotic or late endosomal SNAREs. We conclude that the specificity of SNARE pairing does not suffice to determine the specificity of organelle fusion.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Fasshauer D. Biochim. Biophys. Acta. 2003;1641:87–97. - PubMed
-
- Rizo J., Südhof T. C. Nat. Rev. Neurosci. 2002;3:641–653. - PubMed
-
- Jahn R., Lang T., Südhof T. C. Cell. 2003;112:519–533. - PubMed
-
- Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E. Cell. 1997;90:523–535. - PubMed
-
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Nature. 1998;395:347–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

