BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies
- PMID: 16473845
- PMCID: PMC1363783
- DOI: 10.1093/nar/gnj023
BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies
Abstract
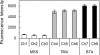
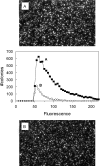
The tricarboxylate reagent benzene-1,3,5-triacetic acid (BTA) was used to attach 5'-aminated DNA primers and templates on an aminosilanized glass surface for subsequent generation of DNA colonies by in situ solid-phase amplification. We have characterized the derivatized surfaces for the chemical attachment of oligonucleotides and evaluate the properties relevant for the amplification process: surface density, thermal stability towards thermocycling, functionalization reproducibility and storage stability. The derivatization process, first developed for glass slides, was then adapted to microfabricated glass channels containing integrated fluidic connections. This implementation resulted in an important reduction of reaction times, consumption of reagents and process automation. Innovative analytical methods for the characterization of attached DNA were developed for assessing the surface immobilized DNA content after amplification. The results obtained showed that the BTA chemistry is compatible and suitable for forming highly dense arrays of DNA colonies with optimal surface coverage of about 10 million colonies/cm(2) from the amplification of initial single-template DNA molecules immobilized. We also demonstrate that the dsDNA colonies generated can be quantitatively processed in situ by restriction enzymes digestion. DNA colonies generated using the BTA reagent can be used for further sequence analysis in an unprecedented parallel fashion for low-cost genomic studies.
Figures






References
-
- Shendure J., Mitra R.D., Varma C., Church G.M. Advanced sequencing technologies: Methods and goals. Nature Rev. Genet. 2004;5:335–344. - PubMed
-
- Carlson C.S., Eberle M.A., Kruglyak L., Nickerson D.A. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. - PubMed
-
- Devlin B., Roeder K., Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 2001;60:155–166. - PubMed
-
- Dobrin S.E., Stephan D.A. Integrating microarrays into disease-gene identification strategies. Expert Rev. Mol. Diagn. 2003;3:375–385. - PubMed
-
- Tebbutt S.J., He J.Q., Burkett K.M., Juan J., Opushnyev I.V., Tripp B.W., Zeznik J.A., Abara C.O., Nelson C.C., Walley K.R. Microarray genotyping resource to determine population stratification in genetic association studies of complex disease. Biotechniques. 2004;37:977–985. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

