Visualizing bacterial tRNA identity determinants and antideterminants using function logos and inverse function logos
- PMID: 16473848
- PMCID: PMC1363773
- DOI: 10.1093/nar/gkj478
Visualizing bacterial tRNA identity determinants and antideterminants using function logos and inverse function logos
Abstract
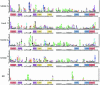
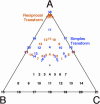
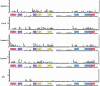
Sequence logos are stacked bar graphs that generalize the notion of consensus sequence. They employ entropy statistics very effectively to display variation in a structural alignment of sequences of a common function, while emphasizing its over-represented features. Yet sequence logos cannot display features that distinguish functional subclasses within a structurally related superfamily nor do they display under-represented features. We introduce two extensions to address these needs: function logos and inverse logos. Function logos display subfunctions that are over-represented among sequences carrying a specific feature. Inverse logos generalize both sequence logos and function logos by displaying under-represented, rather than over-represented, features or functions in structural alignments. To make inverse logos, a compositional inverse is applied to the feature or function frequency distributions before logo construction, where a compositional inverse is a mathematical transform that makes common features or functions rare and vice versa. We applied these methods to a database of structurally aligned bacterial tDNAs to create highly condensed, birds-eye views of potentially all so-called identity determinants and antideterminants that confer specific amino acid charging or initiator function on tRNAs in bacteria. We recovered both known and a few potentially novel identity elements. Function logos and inverse logos are useful tools for exploratory bioinformatic analysis of structure-function relationships in sequence families and superfamilies.
Figures

References
-
- McClain W.H. Transfer RNA identity. FASEB J. 1993;7:72–78. - PubMed
-
- Beuning P.J., Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. - PubMed
-
- Ibba M., Soll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes. Dev. 2004;18:731–738. - PubMed
-
- McClain W.H., Nicholas H.B., Jr Differences between transfer RNA molecules. J. Mol. Biol. 1987;194:635–642. - PubMed

