pH dependence of light-driven proton pumping by an archaerhodopsin from Tibet: comparison with bacteriorhodopsin
- PMID: 16473896
- PMCID: PMC1432102
- DOI: 10.1529/biophysj.105.076547
pH dependence of light-driven proton pumping by an archaerhodopsin from Tibet: comparison with bacteriorhodopsin
Abstract
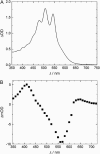
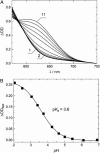
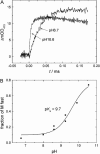
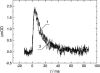
The pH-dependence of photocycle of archaerhodopsin 4 (AR4) was examined, and the underlying proton pumping mechanism investigated. AR4 is a retinal-containing membrane protein isolated from a strain of halobacteria from a Tibetan salt lake. It acts as a light-driven proton pump like bacteriorhodopsin (BR). However, AR4 exhibits an "abnormal" feature--the time sequence of proton release and uptake is reversed at neutral pH. We show here that the temporal sequence of AR4 reversed to "normal"--proton release preceding proton uptake--when the pH is increased above 8.6. We estimated the pK(a) of the proton release complex (PRC) in the M-intermediate to be approximately 8.4, much higher than 5.7 of wide-type BR. The pH-dependence of the rate constant of M-formation shows that the pK(a) of PRC in the initial state of AR4 is approximately 10.4, whereas it is 9.7 in BR. Thus in AR4, the chromophore photoisomerization and subsequent proton transport from the Schiff base to Asp-85 is coupled to a decrease in the pK(a) of PRC from 10.4 to 8.4, which is 2 pK units less than in BR (4 units). This weakened coupling accounts for the lack of early proton release at neutral pH and the reversed time sequence of proton release and uptake in AR4. Nevertheless the PRC in AR4 effectively facilitates deprotonation of primary proton acceptor and recovery of initial state at neutral pH. We found also that all pK(a)s of the key amino acid residues in AR4 were elevated compared to those of BR.
Figures






Similar articles
-
Triton X-100 can alter the temporal sequence of the light-driven proton pump of archaerhodopsin 4.FEBS Lett. 2006 Dec 11;580(28-29):6749-53. doi: 10.1016/j.febslet.2006.11.035. Epub 2006 Nov 28. FEBS Lett. 2006. PMID: 17134701
-
Effect of substitution of proline-77 to aspartate on the light-driven proton release of bacteriorhodopsin.Photochem Photobiol. 2012 Jul-Aug;88(4):922-7. doi: 10.1111/j.1751-1097.2012.01146.x. Epub 2012 Apr 24. Photochem Photobiol. 2012. PMID: 22443335
-
Effects of Triton X-100 on proton transfer and in the photocycle of archaerhodopsin 4.Biosci Biotechnol Biochem. 2012;76(2):250-6. doi: 10.1271/bbb.110508. Epub 2012 Feb 7. Biosci Biotechnol Biochem. 2012. PMID: 22313750
-
Hydration switch model for the proton transfer in the Schiff base region of bacteriorhodopsin.Biochim Biophys Acta. 2004 Jul 23;1658(1-2):72-9. doi: 10.1016/j.bbabio.2004.03.015. Biochim Biophys Acta. 2004. PMID: 15282177 Review.
-
Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport.Biochim Biophys Acta. 2002 Oct 11;1565(2):144-67. doi: 10.1016/s0005-2736(02)00566-7. Biochim Biophys Acta. 2002. PMID: 12409192 Review.
Cited by
-
a-ARM: Automatic Rhodopsin Modeling with Chromophore Cavity Generation, Ionization State Selection, and External Counterion Placement.J Chem Theory Comput. 2019 May 14;15(5):3134-3152. doi: 10.1021/acs.jctc.9b00061. Epub 2019 Apr 12. J Chem Theory Comput. 2019. PMID: 30916955 Free PMC article.
-
Light-Activated Dynamic Clamp Using iPSC-Derived Cardiomyocytes.Biophys J. 2018 Dec 4;115(11):2206-2217. doi: 10.1016/j.bpj.2018.10.018. Epub 2018 Oct 30. Biophys J. 2018. PMID: 30447994 Free PMC article.
-
Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake.Int J Mol Sci. 2022 Jun 21;23(13):6913. doi: 10.3390/ijms23136913. Int J Mol Sci. 2022. PMID: 35805918 Free PMC article.
-
Near-IR resonance Raman spectroscopy of archaerhodopsin 3: effects of transmembrane potential.J Phys Chem B. 2012 Dec 20;116(50):14592-601. doi: 10.1021/jp309996a. Epub 2012 Dec 11. J Phys Chem B. 2012. PMID: 23189985 Free PMC article.
-
Retinal chromophore structure and Schiff base interactions in red-shifted channelrhodopsin-1 from Chlamydomonas augustae.Biochemistry. 2014 Jun 24;53(24):3961-70. doi: 10.1021/bi500445c. Epub 2014 Jun 16. Biochemistry. 2014. PMID: 24869998 Free PMC article.
References
-
- Li, Q. G., Q. G. Sun, W. Zhao, H. Wang, and D. Q. Xu. 2000. Newly isolated archaerhodopsin from a strain of Chinese halobacteria and its proton pumping behavior. Biochim. Biophys. Acta. 1466:260–266. - PubMed
-
- Wang, H., S. X. Zhan, Q. G. Sun, D. Q. Xu, W. Zhao, W. D. Huang, and Q. G. Li. 2000. Primary structure of helix C to helix G of a new retinal protein in H.sp.xz515. Chin. Sci. Bull. 45:1108–1113.
-
- Wang, Y. R., J. Hong, M. Ming, J. D. Ding, Q. G. Li, and W. D. Huang. 2003. Rapid cloning of an archaerhodopsin gene from Halobacterium species xz515 by LPA. J. Fudan University. 42:577–583.
-
- Mukohata, Y. 1994. Comparative-studies on ion pumps of the bacterial rhodopsin family. Biophys. Chem. 50:191–201. - PubMed
-
- Mukohata, Y., Y. Sugiyama, K. Ihara, and M. Yoshida. 1988. An Australian halobacterium contains a novel proton pump retinal protein: archaerhodopsin. Biochem. Biophys. Res. Commun. 151:1339–1345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

