The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis
- PMID: 16473967
- PMCID: PMC1383632
- DOI: 10.1105/tpc.105.039156
The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis
Abstract
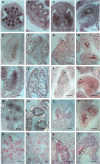
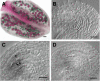
The Arabidopsis-mei2-Like (AML) genes comprise a five-member gene family related to the mei2 gene, which is a master regulator of meiosis in Schizosaccharomyces pombe and encodes an RNA binding protein. We have analyzed the AML genes to assess their role in plant meiosis and development. All five AML genes were expressed in both vegetative and reproductive tissues. Analysis of AML1-AML5 expression at the cellular level indicated a closely similar expression pattern. In the inflorescence, expression was concentrated in the shoot apical meristem, young buds, and reproductive organ primordia. Within the reproductive organs, strong expression was observed in meiocytes and developing gametes. Functional analysis using RNA interference (RNAi) and combinations of insertion alleles revealed a role for the AML genes in meiosis, with RNAi lines and specific multiple mutant combinations displaying sterility and a range of defects in meiotic chromosome behavior. Defects in seedling growth were also observed at low penetrance. These results indicate that the AML genes play a role in meiosis as well as in vegetative growth and reveal conservation in the genetic mechanisms controlling meiosis in yeast and plants.
Figures







References
-
- Agashe, B., Prasad, C.K., and Siddiqi, I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129 3935–3943. - PubMed
-
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Anderson, G.H., Alvarez, N.D., Gilman, C., Jeffares, D.C., Trainor, V.C., Hanson, M.R., and Veit, B. (2004). Diversification of genes encoding mei2-like RNA binding proteins in plants. Plant Mol. Biol. 54 653–670. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

