Retroviral restriction factors Fv1 and TRIM5alpha act independently and can compete for incoming virus before reverse transcription
- PMID: 16474118
- PMCID: PMC1395401
- DOI: 10.1128/JVI.80.5.2100-2105.2006
Retroviral restriction factors Fv1 and TRIM5alpha act independently and can compete for incoming virus before reverse transcription
Abstract
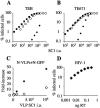
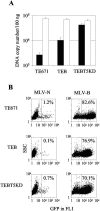
The restriction factors Fv1 and TRIM5alpha provide dominant blocks to retroviral infection, targeting incoming capsids at a postentry, preintegration step. They both restrict N-tropic murine leukemia virus with similar specificity yet act at different points in the viral life cycle. TRIM5alpha-restricted virus is usually unable to reverse transcribe, whereas Fv1-restricted virus reverse transcribes normally. Here we investigate the relationship between these two restriction factors by expressing Fv1 alleles in human cells. We demonstrate that Fv1 is able to compete with TRIM5alpha for virus before reverse transcription. In human cells expressing Fv1(b), N-tropic restricted virus becomes less infectious but reverse transcribes more efficiently, indicating competition between the two antiviral molecules and protection of the virus from TRIM5alpha by Fv1. Our findings suggest that, like TRIM5alpha, Fv1 interacts with virus before reverse transcription, but the consequences of this interaction are not realized until a later stage of the life cycle. We also demonstrate that Fv1 is functionally independent of TRIM5alpha when expressed in human cells.
Figures


Similar articles
-
A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus.Virology. 2008 Sep 1;378(2):233-42. doi: 10.1016/j.virol.2008.05.008. Epub 2008 Jun 30. Virology. 2008. PMID: 18586294 Free PMC article.
-
The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid.J Virol. 2007 Mar;81(5):2138-48. doi: 10.1128/JVI.02318-06. Epub 2006 Nov 29. J Virol. 2007. PMID: 17135314 Free PMC article.
-
Fusion of cyclophilin A to Fv1 enables cyclosporine-sensitive restriction of human and feline immunodeficiency viruses.J Virol. 2007 Sep;81(18):10055-63. doi: 10.1128/JVI.00616-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609268 Free PMC article.
-
Control of viral infectivity by tripartite motif proteins.Hum Gene Ther. 2005 Oct;16(10):1125-32. doi: 10.1089/hum.2005.16.1125. Hum Gene Ther. 2005. PMID: 16218773 Free PMC article. Review.
-
TRIM5alpha.Curr Top Microbiol Immunol. 2009;339:47-66. doi: 10.1007/978-3-642-02175-6_3. Curr Top Microbiol Immunol. 2009. PMID: 20012523 Review.
Cited by
-
Cellular restriction of retrovirus particle-mediated mRNA transfer.J Virol. 2008 Mar;82(6):3069-77. doi: 10.1128/JVI.01880-07. Epub 2008 Jan 16. J Virol. 2008. PMID: 18199655 Free PMC article.
-
Restriction of the felid lentiviruses by a synthetic feline TRIM5-CypA fusion.Vet Immunol Immunopathol. 2011 Oct 15;143(3-4):235-42. doi: 10.1016/j.vetimm.2011.06.017. Epub 2011 Jun 12. Vet Immunol Immunopathol. 2011. PMID: 21813188 Free PMC article. Review.
-
Porcine endogenous retroviruses PERV A and A/C recombinant are insensitive to a range of divergent mammalian TRIM5alpha proteins including human TRIM5alpha.J Gen Virol. 2009 Mar;90(Pt 3):702-709. doi: 10.1099/vir.0.007377-0. J Gen Virol. 2009. PMID: 19218217 Free PMC article.
-
How SAMHD1 changes our view of viral restriction.Trends Immunol. 2012 Jan;33(1):26-33. doi: 10.1016/j.it.2011.11.002. Epub 2011 Dec 15. Trends Immunol. 2012. PMID: 22177690 Free PMC article. Review.
-
Both TRIM5alpha and TRIMCyp have only weak antiviral activity in canine D17 cells.Retrovirology. 2007 Sep 24;4:68. doi: 10.1186/1742-4690-4-68. Retrovirology. 2007. PMID: 17892575 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

