Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1
- PMID: 16474137
- PMCID: PMC1395411
- DOI: 10.1128/JVI.80.5.2309-2317.2006
Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1
Abstract
Herpesviruses replicate their double stranded DNA genomes as high-molecular-weight concatemers which are subsequently cleaved into unit-length genomes by a complex mechanism that is tightly coupled to DNA insertion into a preformed capsid structure, the procapsid. The herpes simplex virus type 1 UL25 protein is incorporated into the capsid during DNA packaging, and previous studies of a null mutant have demonstrated that its function is essential at the late stages of the head-filling process, either to allow packaging to proceed to completion or for retention of the viral genome within the capsid. We have expressed and purified an N-terminally truncated form of the 580-residue UL25 protein and have determined the crystallographic structure of the region corresponding to amino acids 134 to 580 at 2.1-Angstroms resolution. This structure, the first for any herpesvirus protein involved in processing and packaging of viral DNA, reveals a novel fold, a distinctive electrostatic distribution, and a unique "flexible" architecture in which numerous flexible loops emanate from a stable core. Evolutionary trace analysis of UL25 and its homologues in other herpesviruses was used to locate potentially important amino acids on the surface of the protein, leading to the identification of four putative docking regions for protein partners.
Figures

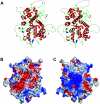
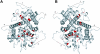
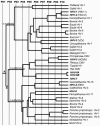

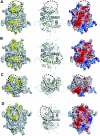
References
-
- Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. - PubMed
-
- Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278-283. - PubMed
-
- Bailey, S. 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. D 50:760-763. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

