Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection
- PMID: 16474153
- PMCID: PMC1395369
- DOI: 10.1128/JVI.80.5.2463-2471.2006
Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection
Abstract
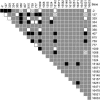
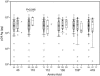
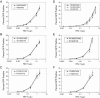
The innate antiviral factor TRIM5alpha restricts the replication of some retroviruses through its interaction with the viral capsid protein, leading to abortive infection. While overexpression of human TRIM5alpha results in modest restriction of human immunodeficiency virus type 1 (HIV-1), this inhibition is insufficient to block productive infection of human cells. We hypothesized that polymorphisms within TRIM5 may result in increased restriction of HIV-1 infection. We sequenced the TRIM5 gene (excluding exon 5) and the 4.8-kb 5' putative regulatory region in genomic DNA from 110 HIV-1-infected subjects and 96 exposed seronegative persons, along with targeted gene sequencing in a further 30 HIV-1-infected individuals. Forty-eight single nucleotide polymorphisms (SNPs), including 20 with allele frequencies of >1.0%, were identified. Among these were two synonymous and eight nonsynonymous coding polymorphisms. We observed no association between TRIM5 polymorphism in HIV-1-infected subjects and their set-point viral load after acute infection, although one TRIM5 haplotype was weakly associated with more rapid CD4(+) T-cell loss. Importantly, a TRIM5 haplotype containing the nonsynonymous SNP R136Q showed increased frequency among HIV-1-infected subjects relative to exposed seronegative persons, with an odds ratio of 5.49 (95% confidence interval = 1.83 to 16.45; P = 0.002). Nonetheless, we observed no effect of individual TRIM5alpha nonsynonymous mutations on the in vitro HIV-1 susceptibility of CD4(+) T cells. Therefore, any effect of TRIM5alpha polymorphism on HIV-1 infection in primary lymphocytes may depend on combinations of SNPs or on DNA sequences in linkage disequilibrium with the TRIM5alpha coding sequence.
Figures


References
-
- An, P., G. Bleiber, P. Duggal, G. Nelson, M. May, B. Mangeat, I. Alobwede, D. Trono, D. Vlahov, S. Donfield, J. J. Goedert, J. Phair, S. Buchbinder, S. J. O'Brien, A. Telenti, and C. A. Winkler. 2004. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 78:11070-11076. - PMC - PubMed
-
- Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. - PubMed
-
- Beyrer, C., A. W. Artenstein, S. Rugpao, H. Stephens, T. C. VanCott, M. L. Robb, M. Rinkaew, D. L. Birx, C. Khamboonruang, P. A. Zimmerman, K. E. Nelson, C. Natpratan, et al. 1999. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. J. Infect. Dis. 179:59-67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

