A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization
- PMID: 16474156
- PMCID: PMC1395368
- DOI: 10.1128/JVI.80.5.2495-2505.2006
A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization
Abstract
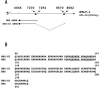
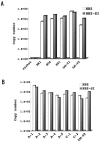
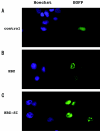
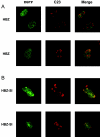
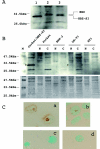
Adult T-cell leukemia (ATL) is associated with prior infection with human T-cell leukemia virus type 1 (HTLV-1); however, the mechanism by which HTLV-1 causes adult T-cell leukemia has not been fully elucidated. Recently, a functional basic leucine zipper (bZIP) protein coded in the minus strand of HTLV-1 genome (HBZ) was identified. We report here a novel isoform of the HTLV-1 bZIP factor (HBZ), HBZ-SI, identified by means of reverse transcription-PCR (RT-PCR) in conjunction with 5' and 3' rapid amplification of cDNA ends (RACE). HBZ-SI is a 206-amino-acid-long protein and is generated by alternative splicing between part of the HBZ gene and a novel exon located in the 3' long terminal repeat of the HTLV-1 genome. Consequently, these isoforms share >95% amino acid sequence identity, and differ only at their N termini, indicating that HBZ-SI is also a functional protein. Duplex RT-PCR and real-time quantitative RT-PCR analyses showed that the mRNAs of these isoforms were expressed at equivalent levels in all ATL cell samples examined. Nonetheless, we found by Western blotting that the HBZ-SI protein was preferentially expressed in some ATL cell lines examined. A key finding was obtained from the subcellular localization analyses of these isoforms. Despite their high sequence similarity, each isoform was targeted to distinguishable subnuclear structures. These data show the presence of a novel isoform of HBZ in ATL cells, and in addition, shed new light on the possibility that each isoform may play a unique role in distinct regions in the cell nucleus.
Figures


References
-
- Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. - PubMed
-
- Basbous, J., C. Arpin, G. Gaudray, M. Piechaczyk, C. Devaux, and J. M. Mesnard. 2003. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278:43620-43627. - PubMed
-
- Calnek, B. W. 1986. Marek's disease—a model for herpesvirus oncology. Crit. Rev. Microbiol. 12:293-320. - PubMed
-
- Cereseto, A., F. Diella, J. C. Mulloy, A. Cara, P. Michieli, R. Grassmann, G. Franchini, and M. E. Klotman. 1996. p53 functional impairment and high p21waf1/cip1 expression in human T-cell lymphotropic/leukemia virus type I-transformed T cells. Blood 88:1551-1560. - PubMed
-
- Furukawa, Y., R. Kubota, M. Tara, S. Izumo, and M. Osame. 2001. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 97:987-993. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

