Crucial role of the 5' conserved structure of bamboo mosaic virus satellite RNA in downregulation of helper viral RNA replication
- PMID: 16474162
- PMCID: PMC1395367
- DOI: 10.1128/JVI.80.5.2566-2574.2006
Crucial role of the 5' conserved structure of bamboo mosaic virus satellite RNA in downregulation of helper viral RNA replication
Erratum in
- J Virol. 2006 Apr;80(8):4204. Annamali, Padmanaban [corrected to Annamalai, Padmanaban]
Abstract
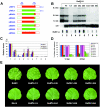
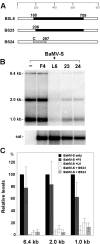
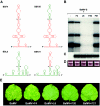
Satellite RNA of Bamboo mosaic virus (satBaMV), a single-stranded mRNA type satellite encoding a protein of 20 kDa (P20), depends on the helper BaMV for replication and encapsidation. Two satBaMV isolates, BSF4 and BSL6, exhibit distinctly differential phenotypes in Nicotiana benthamiana plants when coinoculated with BaMV RNA. BSL6 significantly reduces BaMV RNA replication and suppresses the BaMV-induced symptoms, whereas BSF4 does not. By studies with chimeric satBaMVs generated by exchanging the components between BSF4 and BSL6, the genetic determinants responsible for the downregulation of BaMV replication and symptom expression were mapped at the 5' untranslated region (UTR) of BSL6. The 5' UTR of BSL6 alone is sufficient to diminish BaMV RNA replication when the 5' UTR is inserted in cis into the BaMV expression vector or when coinoculation with mutants that block the synthesis of P20 protein takes place. Further, the 5' UTR of natural satBaMV isolates contains one hypervariable (HV) region which folds into a conserved apical hairpin stem-loop (AHSL) structure (W. B. Yeh, Y. H. Hsu, H. C. Chen, and N. S. Lin, Virology 330:105-115, 2004). Interchanges of AHSL segment of HV regions between BSF4 and BSL6 led to the ability of chimeric satBaMV to interfere with BaMV replication and symptom expression. The conserved secondary structure within the HV region is a potent determinant of the downregulation of helper virus replication.
Figures


References
-
- Annamalai, P., Y. H. Hsu, Y. P. Liu, C. H. Tsai, and N. S. Lin. 2003. Structural and mutational analyses of cis-acting sequences in the 5′-untranslated region of satellite RNA of bamboo mosaic potexvirus. Virology 311:229-239. - PubMed
-
- Baulcombe, D. C., G. R. Saunders, M. W. Bevan, M. A. Mayo, and B. D. Harrison. 1986. Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321:446-449.
-
- Chang, B. Y., N. S. Lin, D. Y. Liou, J. P. Chen, G. G. Liou, and Y. H. Hsu. 1997. Subcellular localization of the 28 kDa protein of the triple-gene-block of bamboo mosaic potexvirus. J. Gen. Virol. 78:1175-1179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

