Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations
- PMID: 16476775
- PMCID: PMC2063674
- DOI: 10.1083/jcb.200507101
Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations
Abstract
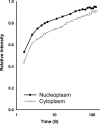
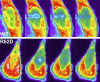
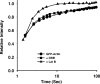
Beta-actin, once thought to be an exclusively cytoplasmic protein, is now known to have important functions within the nucleus. Nuclear beta-actin associates with and functions in chromatin remodeling complexes, ribonucleic acid polymerase complexes, and at least some ribonucleoproteins. Proteins involved in regulating actin polymerization are also found in the interphase nucleus. We define the dynamic properties of nuclear actin molecules using fluorescence recovery after photobleaching. Our results indicate that actin and actin-containing complexes are reduced in their mobility through the nucleoplasm diffusing at approximately 0.5 microm2 s(-1). We also observed that approximately 20% of the total nuclear actin pool has properties of polymeric actin that turns over rapidly. This pool could be detected in endogenous nuclear actin by using fluorescent polymeric actin binding proteins and was sensitive to drugs that alter actin polymerization. Our results validate previous reports of polymeric forms of nuclear actin observed in fixed specimens and reveal that these polymeric forms are very dynamic.
Figures




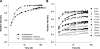

Comment in
-
Nuclear actin: to polymerize or not to polymerize.J Cell Biol. 2006 Feb 13;172(4):495-6. doi: 10.1083/jcb.200601095. J Cell Biol. 2006. PMID: 16476772 Free PMC article. Review.
References
-
- Albuquerque, M.L., and A.S. Flozak. 2001. Patterns of living beta-actin movement in wounded human coronary artery endothelial cells exposed to shear stress. Exp. Cell Res. 270:223–234. - PubMed
-
- Amankwah, K.S., and U. De Boni. 1994. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp. Cell Res. 210:315–325. - PubMed
-
- Andrin, C., and M.J. Hendzel. 2004. F-actin-dependent insolubility of chromatin-modifying components. J. Biol. Chem. 279:25017–25023. - PubMed
-
- Archer, S.K., C. Claudianos, and H.D. Campbell. 2005. Evolution of the gelsolin family of actin-binding proteins as novel transcriptional coactivators. Bioessays. 27:388–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

