Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery
- PMID: 16476776
- PMCID: PMC2063675
- DOI: 10.1083/jcb.200505060
Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery
Abstract
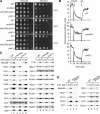
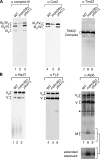
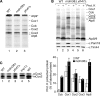
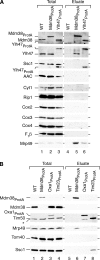
Saccharomyces cerevisiae Mdm38 and Ylh47 are homologues of human Letm1, a protein implicated in Wolf-Hirschhorn syndrome. We analyzed the function of Mdm38 and Ylh47 in yeast mitochondria to gain insight into the role of Letm1. We find that mdm38Delta mitochondria have reduced amounts of certain mitochondrially encoded proteins and low levels of complex III and IV and accumulate unassembled Atp6 of complex V of the respiratory chain. Mdm38 is especially required for efficient transport of Atp6 and cytochrome b across the inner membrane, whereas Ylh47 plays a minor role in this process. Both Mdm38 and Ylh47 form stable complexes with mitochondrial ribosomes, similar to what has been reported for Oxa1, a central component of the mitochondrial export machinery. Our results indicate that Mdm38 functions as a component of an Oxa1-independent insertion machinery in the inner membrane and that Mdm38 plays a critical role in the biogenesis of the respiratory chain by coupling ribosome function to protein transport across the inner membrane.
Figures


References
-
- Chacinska, A., M. Lind, A.E. Frazier, J. Dudek, C. Meisinger, A. Geissler, A. Sickmann, H.E. Meyer, K.N. Truscott, B. Guiard, et al. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120:817–829. - PubMed
-
- DiMauro, S., and E.A. Schon. 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348:2656–2668. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

