Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis
- PMID: 16477027
- PMCID: PMC1413777
- DOI: 10.1073/pnas.0507766103
Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis
Abstract
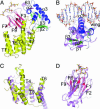
Ser/Thr phosphorylation has emerged as a critical regulatory mechanism in a number of bacteria, including Mycobacterium tuberculosis. This problematic pathogen encodes 11 eukaryotic-like Ser/Thr kinases, yet few substrates or signaling targets have been characterized. Here, we report the structure of EmbR (2.0 A), a putative transcriptional regulator of key arabinosyltransferases (EmbC, -A, and -B), and an endogenous substrate of the Ser/Thr-kinase PknH. EmbR presents a unique domain architecture: the N-terminal winged-helix DNA-binding domain forms an extensive interface with the all-helical central bacterial transcriptional activation domain and is positioned adjacent to the regulatory C-terminal forkhead-associated (FHA) domain, which mediates binding to a Thr-phosphorylated site in PknH. The structure in complex with a phospho-peptide (1.9 A) reveals a conserved mode of phospho-threonine recognition by the FHA domain and evidence for specific recognition of the cognate kinase. The present structures suggest hypotheses as to how EmbR might propagate the phospho-relay signal from its cognate kinase, while serving as a template for the structurally uncharacterized Streptomyces antibiotic regulatory protein family of transcription factors.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Kennelly P. J. FEMS Microbiol. Lett. 2002;206:1–8. - PubMed
-
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., III, et al. Nature. 1998;393:537–544. - PubMed
-
- Av-Gay Y., Everett M. Trends Microbiol. 2000;8:238–244. - PubMed
-
- Duran R., Villarino A., Bellinzoni M., Wehenkel A., Fernandez P., Boitel B., Cole S. T., Alzari P. M., Cervenansky C. Biochem. Biophys. Res. Commun. 2005;333:858–867. - PubMed
-
- Chopra P., Singh B., Singh R., Vohra R., Koul A., Meena L. S., Koduri H., Ghildiyal M., Deol P., Das T. K., et al. Biochem. Biophys. Res. Commun. 2003;311:112–120. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

