Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation
- PMID: 16477312
- PMCID: PMC1361766
- DOI: 10.1371/journal.pgen.0020017
Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation
Abstract
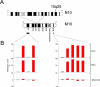
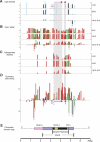
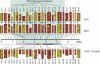
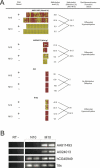
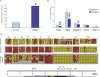
DNA methylation is a hallmark of transcriptional silencing, yet transcription has been reported at the centromere. To address this apparent paradox, we employed a fully sequence-defined ectopic human centromere (or neocentromere) to investigate the relationship between DNA methylation and transcription. We used sodium bisulfite PCR and sequencing to determine the methylation status of 2,041 CpG dinucleotides distributed across a 6.76-Mbp chromosomal region containing a neocentromere. These CpG dinucleotides were associated with conventional and nonconventional CpG islands. We found an overall hypermethylation of the neocentric DNA at nonconventional CpG islands that we designated as CpG islets and CpG orphans. The observed hypermethylation was consistent with the presence of a presumed transcriptionally silent chromatin state at the neocentromere. Within this neocentric chromatin, specific sites of active transcription and the centromeric chromatin boundary are defined by DNA hypomethylation. Our data demonstrate, for the first time to our knowledge, a correlation between DNA methylation and centromere formation in mammals, and that transcription and "chromatin-boundary activity" are permissible at the centromere through the selective hypomethylation of pockets of sequences without compromising the overall silent chromatin state and function of the centromere.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Choo KHA. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. - PubMed
-
- Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. - PubMed
-
- Amor DJ, Kalitsis P, Sumer H, Choo KHA. Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. - PubMed
-
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. - PubMed
-
- Craig JM, Earle E, Canham P, Wong LH, Anderson M, et al. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet. 2003;12:3109–3121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

