Requirement of fission yeast Cid14 in polyadenylation of rRNAs
- PMID: 16478992
- PMCID: PMC1430263
- DOI: 10.1128/MCB.26.5.1710-1721.2006
Requirement of fission yeast Cid14 in polyadenylation of rRNAs
Abstract
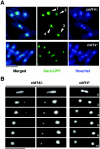
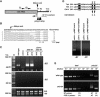
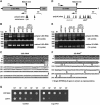
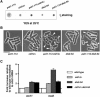
Polyadenylation in eukaryotes is conventionally associated with increased nuclear export, translation, and stability of mRNAs. In contrast, recent studies suggest that the Trf4 and Trf5 proteins, members of a widespread family of noncanonical poly(A) polymerases, share an essential function in Saccharomyces cerevisiae that involves polyadenylation of nuclear RNAs as part of a pathway of exosome-mediated RNA turnover. Substrates for this pathway include aberrantly modified tRNAs and precursors of snoRNAs and rRNAs. Here we show that Cid14 is a Trf4/5 functional homolog in the distantly related fission yeast Schizosaccharomyces pombe. Unlike trf4 trf5 double mutants, cells lacking Cid14 are viable, though they suffer an increased frequency of chromosome missegregation. The Cid14 protein is constitutively nucleolar and is required for normal nucleolar structure. A minor population of polyadenylated rRNAs was identified. These RNAs accumulated in an exosome mutant, and their presence was largely dependent on Cid14, in line with a role for Cid14 in rRNA degradation. Surprisingly, both fully processed 25S rRNA and rRNA processing intermediates appear to be channeled into this pathway. Our data suggest that additional substrates may include the mRNAs of genes involved in meiotic regulation. Polyadenylation-assisted nuclear RNA turnover is therefore likely to be a common eukaryotic mechanism affecting diverse biological processes.
Figures



References
-
- Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Son, Inc., New York, N.Y.
-
- Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641-651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
