Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M
- PMID: 16480273
- PMCID: PMC3015180
- DOI: 10.1021/jm050943c
Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M
Abstract
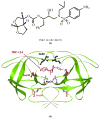
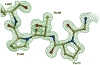
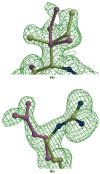
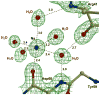
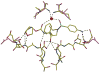
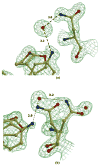
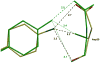
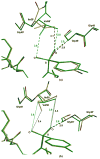
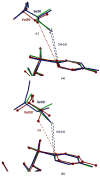
The potent new antiviral inhibitor TMC-114 (UIC-94017) of HIV-1 protease (PR) has been studied with three PR variants containing single mutations D30N, I50V, and L90M, which provide resistance to the major clinical inhibitors. The inhibition constants (K(i)) of TMC-114 for mutants PR(D30N), PR(I50V), and PR(L90M) were 30-, 9-, and 0.14-fold, respectively, relative to wild-type PR. The molecular basis for the inhibition was analyzed using high-resolution (1.22-1.45 A) crystal structures of PR mutant complexes with TMC-114. In PR(D30N), the inhibitor has a water-mediated interaction with the side chain of Asn30 rather than the direct interaction observed in PR, which is consistent with the relative inhibition. Similarly, in PR(I50V) the inhibitor loses favorable hydrophobic interactions with the side chain of Val50. TMC-114 has additional van der Waals contacts in PR(L90M) structure compared to the PR structure, leading to a tighter binding of the inhibitor. The observed changes in PR structure and activity are discussed in relation to the potential for development of resistant mutants on exposure to TMC-114.
Figures
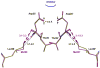
Similar articles
-
Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.J Med Chem. 2007 Sep 6;50(18):4509-15. doi: 10.1021/jm070482q. Epub 2007 Aug 16. J Med Chem. 2007. PMID: 17696515 Free PMC article.
-
Insights into drug resistance of mutations D30N and I50V to HIV-1 protease inhibitor TMC-114: free energy calculation and molecular dynamic simulation.J Mol Model. 2010 Mar;16(3):459-68. doi: 10.1007/s00894-009-0553-7. Epub 2009 Jul 24. J Mol Model. 2010. PMID: 19629548
-
Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.FEBS J. 2005 Oct;272(20):5265-77. doi: 10.1111/j.1742-4658.2005.04923.x. FEBS J. 2005. PMID: 16218957 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients.Antivir Ther. 2004 Jun;9(3):301-14. Antivir Ther. 2004. PMID: 15259893 Review.
Cited by
-
Non-active site mutants of HIV-1 protease influence resistance and sensitisation towards protease inhibitors.Retrovirology. 2020 May 19;17(1):13. doi: 10.1186/s12977-020-00520-6. Retrovirology. 2020. PMID: 32430025 Free PMC article.
-
Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease.Proteins. 2010 Mar;78(4):1015-25. doi: 10.1002/prot.22625. Proteins. 2010. PMID: 19899162 Free PMC article.
-
Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.J Virol. 2007 May;81(10):5144-54. doi: 10.1128/JVI.02706-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360759 Free PMC article.
-
Investigation on the mechanism for the binding and drug resistance of wild type and mutations of G86 residue in HIV-1 protease complexed with Darunavir by molecular dynamic simulation and free energy calculation.J Mol Model. 2014 Feb;20(2):2122. doi: 10.1007/s00894-014-2122-y. Epub 2014 Feb 14. J Mol Model. 2014. PMID: 24526384
-
HIV-1 Protease: Structural Perspectives on Drug Resistance.Viruses. 2009 Dec;1(3):1110-36. doi: 10.3390/v1031110. Epub 2009 Dec 3. Viruses. 2009. PMID: 21994585 Free PMC article.
References
-
- Barlett JA, DeMasi R, Quinn J, Moxham C, Rousseau F. Overview of the Effectiveness of Triple Combination Therapy in Antiretroviral-Naïve HIV-1 Infected Adults. AIDS. 2001;15:1369–1377. - PubMed
-
- Gulick RM, Mellors JW, Havlir D, Eron JJ, Meibohm A, Condra JH, Valentine FT, McMahon D, Gonzalez C, Jonas L, Emini EA, Chodakewitz JA, Isaacs R, Richman DD. 3-Year Suppression if HIV Viremia with Indinavir, Zidovudine, and Lamivudine. Ann Intern Med. 2000;133:35–39. - PubMed
-
- De Clercq E. New Approaches toward Anti-HIV Chemotherapy. J Med Chem. 2005;48:1–17. - PubMed
-
- Menedez-Arias L. Targeting HIV: Antiretroviral Therapy and Development of Drug Resistance. Trends Pharmacol Sci. 2002;23:381–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials

