BIOZON: a system for unification, management and analysis of heterogeneous biological data
- PMID: 16480510
- PMCID: PMC1449871
- DOI: 10.1186/1471-2105-7-70
BIOZON: a system for unification, management and analysis of heterogeneous biological data
Abstract
Background: Integration of heterogeneous data types is a challenging problem, especially in biology, where the number of databases and data types increase rapidly. Amongst the problems that one has to face are integrity, consistency, redundancy, connectivity, expressiveness and updatability.
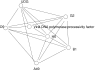
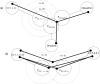
Description: Here we present a system (Biozon) that addresses these problems, and offers biologists a new knowledge resource to navigate through and explore. Biozon unifies multiple biological databases consisting of a variety of data types (such as DNA sequences, proteins, interactions and cellular pathways). It is fundamentally different from previous efforts as it uses a single extensive and tightly connected graph schema wrapped with hierarchical ontology of documents and relations. Beyond warehousing existing data, Biozon computes and stores novel derived data, such as similarity relationships and functional predictions. The integration of similarity data allows propagation of knowledge through inference and fuzzy searches. Sophisticated methods of query that span multiple data types were implemented and first-of-a-kind biological ranking systems were explored and integrated.
Conclusion: The Biozon system is an extensive knowledge resource of heterogeneous biological data. Currently, it holds more than 100 million biological documents and 6.5 billion relations between them. The database is accessible through an advanced web interface that supports complex queries, "fuzzy" searches, data materialization and more, online at http://biozon.org.
Figures



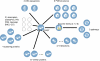
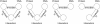


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

