The expansion of the metazoan microRNA repertoire
- PMID: 16480513
- PMCID: PMC1388199
- DOI: 10.1186/1471-2164-7-25
The expansion of the metazoan microRNA repertoire
Abstract
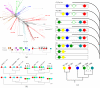
Background: MicroRNAs have been identified as crucial regulators in both animals and plants. Here we report on a comprehensive comparative study of all known miRNA families in animals. We expand the MicroRNA Registry 6.0 by more than 1000 new homologs of miRNA precursors whose expression has been verified in at least one species. Using this uniform data basis we analyze their evolutionary history in terms of individual gene phylogenies and in terms of preservation of genomic nearness across species. This allows us to reliably identify microRNA clusters that are derived from a common transcript.
Results: We identify three episodes of microRNA innovation that correspond to major developmental innovations: A class of about 20 miRNAs is common to protostomes and deuterostomes and might be related to the advent of bilaterians. A second large wave of innovations maps to the branch leading to the vertebrates. The third significant outburst of miRNA innovation coincides with placental (eutherian) mammals. In addition, we observe the expected expansion of the microRNA inventory due to genome duplications in early vertebrates and in an ancestral teleost. The non-local duplications in the vertebrate ancestor are predated by local (tandem) duplications leading to the formation of about a dozen ancient microRNA clusters.
Conclusion: Our results suggest that microRNA innovation is an ongoing process. Major expansions of the metazoan miRNA repertoire coincide with the advent of bilaterians, vertebrates, and (placental) mammals.
Figures





Similar articles
-
Evolution of the let-7 microRNA family.RNA Biol. 2012 Mar;9(3):231-41. doi: 10.4161/rna.18974. Epub 2012 Mar 1. RNA Biol. 2012. PMID: 22617875 Free PMC article.
-
Simultaneous expansions of microRNAs and protein-coding genes by gene/genome duplications in early vertebrates.J Exp Zool B Mol Dev Evol. 2009 May 15;312B(3):164-70. doi: 10.1002/jez.b.21273. J Exp Zool B Mol Dev Evol. 2009. PMID: 19214983
-
MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes.PLoS One. 2011 Mar 16;6(3):e17666. doi: 10.1371/journal.pone.0017666. PLoS One. 2011. PMID: 21436881 Free PMC article.
-
Evolution of microRNAs.Methods Mol Biol. 2006;342:335-50. doi: 10.1385/1-59745-123-1:335. Methods Mol Biol. 2006. PMID: 16957387 Review.
-
Evolution of plant microRNA gene families.Cell Res. 2007 Mar;17(3):212-8. doi: 10.1038/sj.cr.7310113. Cell Res. 2007. PMID: 17130846 Review.
Cited by
-
microRNAs and the evolution of complex multicellularity: identification of a large, diverse complement of microRNAs in the brown alga Ectocarpus.Nucleic Acids Res. 2015 Jul 27;43(13):6384-98. doi: 10.1093/nar/gkv578. Epub 2015 Jun 22. Nucleic Acids Res. 2015. PMID: 26101255 Free PMC article.
-
On the Nature and Evolutionary Impact of Phenotypic Robustness Mechanisms.Annu Rev Ecol Evol Syst. 2014 Nov 1;45:496-517. doi: 10.1146/annurev-ecolsys-120213-091705. Annu Rev Ecol Evol Syst. 2014. PMID: 26034410 Free PMC article.
-
Evolution of the let-7 microRNA family.RNA Biol. 2012 Mar;9(3):231-41. doi: 10.4161/rna.18974. Epub 2012 Mar 1. RNA Biol. 2012. PMID: 22617875 Free PMC article.
-
Clusters of microRNAs emerge by new hairpins in existing transcripts.Nucleic Acids Res. 2013 Sep;41(16):7745-52. doi: 10.1093/nar/gkt534. Epub 2013 Jun 17. Nucleic Acids Res. 2013. PMID: 23775791 Free PMC article.
-
Characterization and evolution of microRNA genes derived from repetitive elements and duplication events in plants.PLoS One. 2012;7(4):e34092. doi: 10.1371/journal.pone.0034092. Epub 2012 Apr 16. PLoS One. 2012. PMID: 22523544 Free PMC article.
References
-
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kurodak MI, Mailer B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

