Evolution of antibody class switching: identification and transcriptional control of an Inu exon in the duck (Anas platyrhynchos)
- PMID: 16480768
- PMCID: PMC1400595
- DOI: 10.1016/j.dci.2005.05.004
Evolution of antibody class switching: identification and transcriptional control of an Inu exon in the duck (Anas platyrhynchos)
Abstract
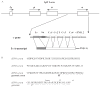
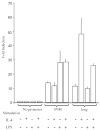
Immunoglobulin class switching is characteristic to the tetrapod lineage, but the nature of this process has been elucidated only in mammals, where I-exon transcription initiates and directs the recombination in the IgH locus. Here, it is shown that an I-exon occurs 5' of the nu (IgY constant region) gene of the duck (Anas platyrhynchos): it is longer than mammalian I-exons and comprised primarily of tandem repeats. The Inu promoter was identified and shown to be responsive to stimulation with IL-4 but not LPS. It contains Oct, LYF-1, ATF, and C/EBP motifs. Site directed mutagenesis indicates that 2 C/EBP motifs are uniquely necessary for the response of the promoter to IL-4, as tested in the mouse pre-B cell line, 70Z/3. These results support the conclusion that the signal transduction pathways controlling I-exon promoter responses to cytokines have been highly conserved in vertebrate evolution.
Figures
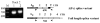




Similar articles
-
Enhancer and promoter activity in the JH to IGHM intron of the Pekin duck, Anas platyrhynchos.Dev Comp Immunol. 2007;31(3):286-95. doi: 10.1016/j.dci.2006.06.004. Epub 2006 Aug 4. Dev Comp Immunol. 2007. PMID: 16930702
-
One gene encodes the heavy chains for three different forms of IgY in the duck.J Immunol. 1994 Dec 15;153(12):5549-55. J Immunol. 1994. PMID: 7989756
-
Structural relationship between the two IgY of the duck, Anas platyrhynchos: molecular genetic evidence.J Immunol. 1992 Oct 15;149(8):2627-33. J Immunol. 1992. PMID: 1401901
-
The molecular events in heavy chain class-switching.Semin Immunol. 1989 Sep;1(1):65-77. Semin Immunol. 1989. PMID: 15630960 Review.
-
Immunoglobulin class switching.Curr Opin Immunol. 1996 Apr;8(2):199-205. doi: 10.1016/s0952-7915(96)80058-6. Curr Opin Immunol. 1996. PMID: 8725943 Review.
References
-
- Stavnezer J, Kinoshito K, Muramatsu M, Honjo T. Molecular mechanism of class switch recombination. In: Honjo T, Alt FW, Neuberger M, editors. Molecular Biology of B Cells. London: Elsevier Academic Press; 2004. p. 307–26.
-
- Kinoshita K, Lee CG, Tashiro J, Muramatsu M, Chen XC, Yoshikawa K, et al. Molecular mechanism of immunoglobulin class switch recombination. Cold Spring Harb Symp Quant Biol. 1999;64:217–26. - PubMed
-
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274(26):18,470–18,476. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

