Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites
- PMID: 16481621
- PMCID: PMC1413848
- DOI: 10.1073/pnas.0511216103
Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites
Abstract
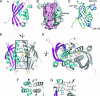
Bacterial type IV secretion systems (T4SS) translocate DNA and/or proteins to recipient cells, thus providing a mechanism for conjugative transfer of genetic material and bacterial pathogenesis. Here we describe the first structure of a core component from the archetypal Agrobacterium tumefaciens T4SS: the 2.2-A resolution crystal structure of the VirB8 periplasmic domain (pVirB8(AT)). VirB8 forms a dimer in the crystal, and we identify residues likely important for stabilization of the dimer interface. Structural comparison of pVirB8(AT) with Brucella suis VirB8 confirms that the monomers have a similar fold. In addition, the pVirB8(AT) dimer superimposes very closely on the B. suis VirB8 dimer, supporting the proposal that dimer formation in the crystal reflects self-interactions that are biologically significant. The evolutionary conservation level for each residue was obtained from a data set of 84 VirB8 homologs and projected onto the protein structure to indicate conserved surface patches that likely contact other T4SS proteins.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

