Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays
- PMID: 16481660
- PMCID: PMC1457032
- DOI: 10.1101/gr.4337206
Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays
Abstract
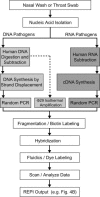
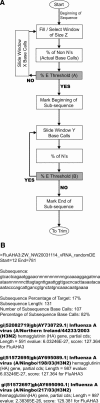
The exponential growth of pathogen nucleic acid sequences available in public domain databases has invited their direct use in pathogen detection, identification, and surveillance strategies. DNA microarray technology has offered the potential for the direct DNA sequence analysis of a broad spectrum of pathogens of interest. However, to achieve the practical attainment of this potential, numerous technical issues, especially nucleic acid amplification, probe specificity, and interpretation strategies of sequence detection, need to be addressed. In this report, we demonstrate an approach that combines the use of a custom-designed Affymetrix resequencing Respiratory Pathogen Microarray (RPM v.1) with methods for microbial nucleic acid enrichment, random nucleic acid amplification, and automated sequence similarity searching for broad-spectrum respiratory pathogen surveillance. Successful proof-of-concept experiments, utilizing clinical samples obtained from patients presenting adenovirus or influenza virus-induced febrile respiratory illness (FRI), demonstrate the ability of this approach for correct species- and strain-level identification with unambiguous statistical interpretation at clinically relevant sensitivity levels. Our results underscore the feasibility of using this approach to expedite the early surveillance of diseases, and provide new information on the incidence of multiple pathogens.
Figures


References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Bodrossy L., Sessitsch A., Sessitsch A. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 2004;7:1–10. - PubMed
-
- Boivin G., Coulombe Z., Wat C., Coulombe Z., Wat C., Wat C. Quantification of the influenza virus load by real-time polymerase chain reaction in nasopharyngeal swabs of patients treated with oseltamivir. J. Infect. Dis. 2003;188:578–580. - PubMed
-
- Bryant P.A., Venter D., Robins-Browne R., Curtis N., Venter D., Robins-Browne R., Curtis N., Robins-Browne R., Curtis N., Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect. Dis. 2004;4:100–111. - PubMed
-
- Cleland C.A., White P.S., Deshpande A., Wolinsky M., Song J., Nolan J.P., White P.S., Deshpande A., Wolinsky M., Song J., Nolan J.P., Deshpande A., Wolinsky M., Song J., Nolan J.P., Wolinsky M., Song J., Nolan J.P., Song J., Nolan J.P., Nolan J.P. Development of rationally designed nucleic acid signatures for microbial pathogens. Expert Rev. Mol. Diagn. 2004;4:303–315. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
