Bias of selection on human copy-number variants
- PMID: 16482228
- PMCID: PMC1366494
- DOI: 10.1371/journal.pgen.0020020
Bias of selection on human copy-number variants
Abstract
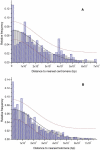
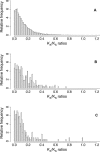
Although large-scale copy-number variation is an important contributor to conspecific genomic diversity, whether these variants frequently contribute to human phenotype differences remains unknown. If they have few functional consequences, then copy-number variants (CNVs) might be expected both to be distributed uniformly throughout the human genome and to encode genes that are characteristic of the genome as a whole. We find that human CNVs are significantly overrepresented close to telomeres and centromeres and in simple tandem repeat sequences. Additionally, human CNVs were observed to be unusually enriched in those protein-coding genes that have experienced significantly elevated synonymous and nonsynonymous nucleotide substitution rates, estimated between single human and mouse orthologues. CNV genes encode disproportionately large numbers of secreted, olfactory, and immunity proteins, although they contain fewer than expected genes associated with Mendelian disease. Despite mouse CNVs also exhibiting a significant elevation in synonymous substitution rates, in most other respects they do not differ significantly from the genomic background. Nevertheless, they encode proteins that are depleted in olfactory function, and they exhibit significantly decreased amino acid sequence divergence. Natural selection appears to have acted discriminately among human CNV genes. The significant overabundance, within human CNVs, of genes associated with olfaction, immunity, protein secretion, and elevated coding sequence divergence, indicates that a subset may have been retained in the human population due to the adaptive benefit of increased gene dosage. By contrast, the functional characteristics of mouse CNVs either suggest that advantageous gene copies have been depleted during recent selective breeding of laboratory mouse strains or suggest that they were preferentially fixed as a consequence of the larger effective population size of wild mice. It thus appears that CNV differences among mouse strains do not provide an appropriate model for large-scale sequence variations in the human population.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
-
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

