Proteomic analysis of altered protein expression in skeletal muscle of rats in a hypermetabolic state induced by burn sepsis
- PMID: 16483253
- PMCID: PMC1479762
- DOI: 10.1042/BJ20051710
Proteomic analysis of altered protein expression in skeletal muscle of rats in a hypermetabolic state induced by burn sepsis
Abstract
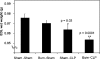
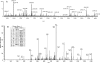
mRNA profiling has been extensively used to study muscle wasting. mRNA level changes may not reflect that of proteins, especially in catabolic muscle where there is decreased synthesis and increased degradation. As sepsis is often associated with burn injury, and burn superimposed by sepsis has been shown to result in significant loss of lean tissues, we characterized changes in the skeletal-muscle proteome of rats subjected to a cutaneous burn covering 20% of the total body surface area, followed 2 days later by sepsis induced by CLP (caecal ligation and puncture). EDL (extensor digitorum longus) muscles were dissected from Burn-CLP animals (n=4) and controls (sham-burned and sham-CLP-treated, n=4). Burn-CLP injury resulted in a rapid loss of EDL weight, increased ubiquitin-conjugated proteins and increased protein carbonyl groups. EDL protein profiles were obtained by two-dimensional gel electrophoresis using two immobilized pH gradient strips with overlapping pH range covering a pH 3-8 range. Seventeen spots were significantly altered in the Burn-CLP compared with the control group, representing 15 different proteins identified by peptide mass fingerprinting. The identities of three proteins including transferrin were further confirmed by liquid chromatography-tandem MS. The significant changes in transferrin and HSP27 (heat-shock protein 27) were verified by Western-blot analysis. HSP60, HSP27 and HSPbeta6 were down-regulated, along with HSP70, as detected by Western blotting. Six metabolic enzymes related to energy production were also down-regulated. A simultaneous decrease in chaperone proteins and metabolic enzymes could decrease protein synthesis. Furthermore, decreased HSPs could increase oxidative damage, thus accelerating protein degradation. Using cultured C2C12 myotubes, we showed that H2O2-induced protein degradation in vitro could be partially attenuated by prior heat-shock treatment, consistent with a protective role of HSP70 and/or other HSPs against proteolysis.
Figures







Similar articles
-
Contribution of gene expression to metabolic fluxes in hypermetabolic livers induced through burn injury and cecal ligation and puncture in rats.Biotechnol Bioeng. 2007 May 1;97(1):118-37. doi: 10.1002/bit.21200. Biotechnol Bioeng. 2007. PMID: 17009336 Free PMC article.
-
[Changes in skeletal muscle protein metabolism in burned rats with sepsis and the role of glucocorticoid in skeletal muscle proteolysis].Zhonghua Wai Ke Za Zhi. 2002 Sep;40(9):705-8. Zhonghua Wai Ke Za Zhi. 2002. PMID: 12411147 Chinese.
-
Sepsis and glucocorticoids downregulate the expression of the nuclear cofactor PGC-1beta in skeletal muscle.Am J Physiol Endocrinol Metab. 2010 Oct;299(4):E533-43. doi: 10.1152/ajpendo.00596.2009. Epub 2010 Jul 20. Am J Physiol Endocrinol Metab. 2010. PMID: 20647557 Free PMC article.
-
Burn-induced hypermetabolism and skeletal muscle dysfunction.Am J Physiol Cell Physiol. 2021 Jul 1;321(1):C58-C71. doi: 10.1152/ajpcell.00106.2021. Epub 2021 Apr 28. Am J Physiol Cell Physiol. 2021. PMID: 33909503 Free PMC article. Review.
-
Muscle in defense.Crit Care Med. 2009 Oct;37(10 Suppl):S384-90. doi: 10.1097/CCM.0b013e3181b6f8a5. Crit Care Med. 2009. PMID: 20046124 Review.
Cited by
-
Roles and potential therapeutic targets of the ubiquitin proteasome system in muscle wasting.BMC Biochem. 2007 Nov 22;8 Suppl 1(Suppl 1):S7. doi: 10.1186/1471-2091-8-S1-S7. BMC Biochem. 2007. PMID: 18047744 Free PMC article. Review.
-
Proteolysis activation and proteome alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension.Eur J Appl Physiol. 2009 Nov;107(5):553-63. doi: 10.1007/s00421-009-1151-1. Epub 2009 Aug 19. Eur J Appl Physiol. 2009. PMID: 19690883
-
Antiapoptotic effect of a novel synthetic peptide from bovine muscle and MPG peptide on H2O2-induced C2C12 cells.In Vitro Cell Dev Biol Anim. 2014 Aug;50(7):630-9. doi: 10.1007/s11626-014-9745-2. Epub 2014 May 14. In Vitro Cell Dev Biol Anim. 2014. PMID: 24825375
-
SILAC analysis of oxidative stress-mediated proteins in human pneumocytes: new role for treacle.Proteomics. 2010 Jun;10(11):2165-74. doi: 10.1002/pmic.201000020. Proteomics. 2010. PMID: 20340163 Free PMC article.
-
Temporal study following burn injury in young rats is associated with skeletal muscle atrophy, inflammation and altered myogenic regulatory factors.Inflamm Res. 2015 Jan;64(1):53-62. doi: 10.1007/s00011-014-0783-8. Epub 2014 Nov 21. Inflamm Res. 2015. PMID: 25413930
References
-
- Lecker S. H., Solomon V., Mitch W. E., Goldberg A. L. Muscle protein breakdown and the critical role of the ubiquitin–proteasome pathway in normal and disease states. J. Nutr. 1999;129:227S–237S. - PubMed
-
- Hasselgren P. O. Catabolic response to stress and injury: implications for regulation. World J. Surg. 2000;24:1452–1459. - PubMed
-
- Hobler S. C., Tiao G., Fischer J. E., Monaco J., Hasselgren P. O. Sepsis-induced increase in muscle proteolysis is blocked by specific proteasome inhibitors. Am. J. Physiol. 1998;274:R30–R37. - PubMed
-
- Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

