Tracking the evolution of alternatively spliced exons within the Dscam family
- PMID: 16483367
- PMCID: PMC1397879
- DOI: 10.1186/1471-2148-6-16
Tracking the evolution of alternatively spliced exons within the Dscam family
Abstract
Background: The Dscam gene in the fruit fly, Drosophila melanogaster, contains twenty-four exons, four of which are composed of tandem arrays that each undergo mutually exclusive alternative splicing (4, 6, 9 and 17), potentially generating 38,016 protein isoforms. This degree of transcript diversity has not been found in mammalian homologs of Dscam. We examined the molecular evolution of exons within this gene family to locate the point of divergence for this alternative splicing pattern.
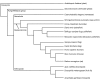
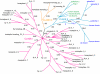
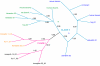
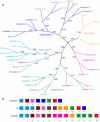
Results: Using the fruit fly Dscam exons 4, 6, 9 and 17 as seed sequences, we iteratively searched sixteen genomes for homologs, and then performed phylogenetic analyses of the resulting sequences to examine their evolutionary history. We found homologs in the nematode, arthropod and vertebrate genomes, including homologs in several vertebrates where Dscam had not been previously annotated. Among these, only the arthropods contain homologs arranged in tandem arrays indicative of mutually exclusive splicing. We found no homologs to these exons within the Arabidopsis, yeast, tunicate or sea urchin genomes but homologs to several constitutive exons from fly Dscam were present within tunicate and sea urchin. Comparing the rate of turnover within the tandem arrays of the insect taxa (fruit fly, mosquito and honeybee), we found the variants within exons 4 and 17 are well conserved in number and spatial arrangement despite 248-283 million years of divergence. In contrast, the variants within exons 6 and 9 have undergone considerable turnover since these taxa diverged, as indicated by deeply branching taxon-specific lineages.
Conclusion: Our results suggest that at least one Dscam exon array may be an ancient duplication that predates the divergence of deuterostomes from protostomes but that there is no evidence for the presence of arrays in the common ancestor of vertebrates. The different patterns of conservation and turnover among the Dscam exon arrays provide a striking example of how a gene can evolve in a modular fashion rather than as a single unit.
Figures



Similar articles
-
More than one way to produce protein diversity: duplication and limited alternative splicing of an adhesion molecule gene in basal arthropods.Evolution. 2013 Oct;67(10):2999-3011. doi: 10.1111/evo.12179. Epub 2013 Jul 4. Evolution. 2013. PMID: 24094349
-
The evolution of Dscam genes across the arthropods.BMC Evol Biol. 2012 Apr 13;12:53. doi: 10.1186/1471-2148-12-53. BMC Evol Biol. 2012. PMID: 22500922 Free PMC article.
-
The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects.Mol Biol Evol. 2008 Jul;25(7):1429-39. doi: 10.1093/molbev/msn087. Epub 2008 Apr 9. Mol Biol Evol. 2008. PMID: 18403399
-
Evolutionary convergence of alternative splicing in ion channels.Trends Genet. 2004 Apr;20(4):171-6. doi: 10.1016/j.tig.2004.02.001. Trends Genet. 2004. PMID: 15101391 Review.
-
Alternative splicing of mutually exclusive exons--a review.Biosystems. 2013 Oct;114(1):31-8. doi: 10.1016/j.biosystems.2013.07.003. Epub 2013 Jul 11. Biosystems. 2013. PMID: 23850531 Review.
Cited by
-
From Biomphalaria glabrata to Drosophila melanogaster and Anopheles gambiae: the diversity and role of FREPs and Dscams in immune response.Front Immunol. 2025 Apr 30;16:1579905. doi: 10.3389/fimmu.2025.1579905. eCollection 2025. Front Immunol. 2025. PMID: 40370466 Free PMC article. Review.
-
AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system.PLoS Biol. 2006 Jul;4(7):e229. doi: 10.1371/journal.pbio.0040229. PLoS Biol. 2006. PMID: 16774454 Free PMC article.
-
Down syndrome cell adhesion molecule 1: testing for a role in insect immunity, behaviour and reproduction.R Soc Open Sci. 2016 Apr 20;3(4):160138. doi: 10.1098/rsos.160138. eCollection 2016 Apr. R Soc Open Sci. 2016. PMID: 27152227 Free PMC article.
-
Exploring perspectives of Dscam for cognitive deficits: a review of multifunction for regulating neural wiring in homeostasis.Front Mol Neurosci. 2025 May 2;18:1575348. doi: 10.3389/fnmol.2025.1575348. eCollection 2025. Front Mol Neurosci. 2025. PMID: 40385132 Free PMC article. Review.
-
Expansion of stochastic expression repertoire by tandem duplication in mouse Protocadherin-α cluster.Sci Rep. 2014 Sep 2;4:6263. doi: 10.1038/srep06263. Sci Rep. 2014. PMID: 25179445 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

