The in vitro real-time oscillation monitoring system identifies potential entrainment factors for circadian clocks
- PMID: 16483373
- PMCID: PMC1386696
- DOI: 10.1186/1471-2199-7-5
The in vitro real-time oscillation monitoring system identifies potential entrainment factors for circadian clocks
Abstract
Background: Circadian rhythms are endogenous, self-sustained oscillations with approximately 24-hr rhythmicity that are manifested in various physiological and metabolic processes. The circadian organization of these processes in mammals is governed by the master oscillator within the suprachiasmatic nuclei (SCN) of the hypothalamus. Recent findings revealed that circadian oscillators exist in most organs, tissues, and even in immortalized cells, and that the oscillators in peripheral tissues are likely to be coordinated by SCN, the master oscillator. Some candidates for endogenous entrainment factors have sporadically been reported, however, their details remain mainly obscure.
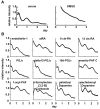
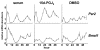
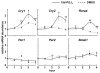
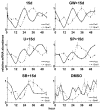
Results: We developed the in vitro real-time oscillation monitoring system (IV-ROMS) by measuring the activity of luciferase coupled to the oscillatory gene promoter using photomultiplier tubes and applied this system to screen and identify factors able to influence circadian rhythmicity. Using this IV-ROMS as the primary screening of entrainment factors for circadian clocks, we identified 12 candidates as the potential entrainment factor in a total of 299 peptides and bioactive lipids. Among them, four candidates (endothelin-1, all-trans retinoic acid, 9-cis retinoic acid, and 13-cis retinoic acid) have already been reported as the entrainment factors in vivo and in vitro. We demonstrated that one of the novel candidates, 15-deoxy-Delta12,14-prostaglandin J2 (15d-PGJ2), a natural ligand of the peroxisome proliferator-activated receptor-gamma (PPAR-gamma), triggers the rhythmic expression of endogenous clock genes in NIH3T3 cells. Furthermore, we showed that 15d-PGJ2 transiently induces Cry1, Cry2, and Roralpha mRNA expressions and that 15d-PGJ2-induced entrainment signaling pathway is PPAR-gamma--and MAPKs (ERK, JNK, p38MAPK)-independent.
Conclusion: Here, we identified 15d-PGJ2 as an entrainment factor in vitro. Using our developed IV-ROMS to screen 299 compounds, we found eight novel and four known molecules to be potential entrainment factors for circadian clocks, indicating that this assay system is a powerful and useful tool in initial screenings.
Figures
Similar articles
-
Circadian entrainment aftereffects in suprachiasmatic nuclei and peripheral tissues in vitro.Brain Res. 2008 Sep 4;1228:127-34. doi: 10.1016/j.brainres.2008.05.091. Epub 2008 Jun 14. Brain Res. 2008. PMID: 18598681
-
Transcriptional oscillation of canonical clock genes in mouse peripheral tissues.BMC Mol Biol. 2004 Oct 9;5:18. doi: 10.1186/1471-2199-5-18. BMC Mol Biol. 2004. PMID: 15473909 Free PMC article.
-
The rat circadian clockwork and its photoperiodic entrainment during development.Chronobiol Int. 2006;23(1-2):237-43. doi: 10.1080/07420520500522523. Chronobiol Int. 2006. PMID: 16687297
-
Properties, entrainment, and physiological functions of mammalian peripheral oscillators.J Biol Rhythms. 2006 Dec;21(6):494-506. doi: 10.1177/0748730406293889. J Biol Rhythms. 2006. PMID: 17107939 Review.
-
Physiology of circadian entrainment.Physiol Rev. 2010 Jul;90(3):1063-102. doi: 10.1152/physrev.00009.2009. Physiol Rev. 2010. PMID: 20664079 Review.
Cited by
-
Improved PCR methods for detection of African rabies and rabies-related lyssaviruses.J Clin Microbiol. 2010 Nov;48(11):3949-55. doi: 10.1128/JCM.01256-10. Epub 2010 Sep 1. J Clin Microbiol. 2010. PMID: 20810772 Free PMC article.
-
Progesterone, but not estradiol, synchronizes circadian oscillator in the uterus endometrial stromal cells.Mol Cell Biochem. 2009 Apr;324(1-2):31-8. doi: 10.1007/s11010-008-9981-4. Epub 2008 Dec 20. Mol Cell Biochem. 2009. PMID: 19096762
-
Lipocalin-type prostaglandin D synthase regulates light-induced phase advance of the central circadian rhythm in mice.Commun Biol. 2020 Oct 8;3(1):557. doi: 10.1038/s42003-020-01281-w. Commun Biol. 2020. PMID: 33033338 Free PMC article.
-
Retinoic Acid Signalling in the Pineal Gland Is Conserved across Mammalian Species and Its Transcriptional Activity Is Inhibited by Melatonin.Cells. 2023 Jan 11;12(2):286. doi: 10.3390/cells12020286. Cells. 2023. PMID: 36672220 Free PMC article.
-
Resolvin D1 and resolvin D2 govern local inflammatory tone in obese fat.J Immunol. 2012 Sep 1;189(5):2597-605. doi: 10.4049/jimmunol.1201272. Epub 2012 Jul 27. J Immunol. 2012. PMID: 22844113 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

