Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP)
- PMID: 16483380
- PMCID: PMC1434769
- DOI: 10.1186/1471-2156-7-10
Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP)
Abstract
Background: Sesame is an important oil crop in tropical and subtropical areas. Despite its nutritional value and historic and cultural importance, the research on sesame has been scarce, particularly as far as its genetic diversity is concerned. The aims of the present study were to clarify genetic relationships among 32 sesame accessions from the Venezuelan Germplasm Collection, which represents genotypes from five diversity centres (India, Africa, China-Korea-Japan, Central Asia and Western Asia), and to determine the association between geographical origin and genetic diversity using amplified fragment length polymorphism (AFLP).
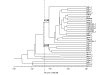
Results: Large genetic variability was found within the germplasm collection. A total of 457 AFLP markers were recorded, 93 % of them being polymorphic. The Jaccard similarity coefficient ranged from 0.38 to 0.85 between pairs of accessions. The UPGMA dendrogram grouped 25 of 32 accessions in two robust clusters, but it has not revealed any association between genotype and geographical origin. Indian, African and Chinese-Korean-Japanese accessions were distributed throughout the dendrogram. A similar pattern was obtained using principal coordinates analysis. Genetic diversity studies considering five groups of accessions according to the geographic origin detected that only 20 % of the total diversity was due to diversity among groups using Nei's coefficient of population differentiation. Similarly, only 5% of the total diversity was attributed to differences among groups by the analysis of molecular variance (AMOVA). This small but significant difference was explained by the fact that the Central Asia group had a lower genetic variation than the other diversity centres studied.
Conclusion: We found that our sesame collection was genetically very variable and did not show an association between geographical origin and AFLP patterns. This result suggests that there was considerable gene flow among diversity centres. Future germplasm collection strategies should focus on sampling a large number of plants. Covering many diversity centres is less important because each centre represents a major part of the total diversity in sesame, Central Asia centre being the only exception. The same recommendation holds for the choice of parents for segregant populations used in breeding projects. The traditional assumption that selecting genotypes of different geographical origin will maximize the diversity available to a breeding project does not hold in sesame.
Figures

References
-
- Bedigian D, Harlan J. Evidence for cultivation on sesame in the ancient world. Economic Botany. 1986;40:137–154.
-
- Ashri A. Sesame breeding. Plant Breeding Reviews. 1998;16:179–228.
-
- FAO FAOstat Databases. 2005. http://faostat.fao.org/
-
- Brar G, Ahuja R. Sesame: its culture, genetics, breeding and biochemistry. In: Malik CP, editor. Annu Rev Plant Sci. New Dehli, Kalyani publishers; 1979. pp. 285–313.
-
- Bedigian D. Evolution of sesame revisited: domestication, diversity and prospects. Genetic Resources and Crop Evolution. 2003;50:779–787.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

