Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori
- PMID: 16484186
- PMCID: PMC1426556
- DOI: 10.1128/JB.188.5.1750-1761.2006
Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori
Abstract
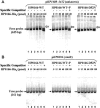
About 200 genes of the gastric pathogen Helicobacter pylori increase expression at medium pHs of 6.2, 5.5, and 4.5, an increase that is abolished or much reduced by the buffering action of urease. Genes up-regulated by a low pH include the two-component system HP0165-HP0166, suggesting a role in the regulation of some of the pH-sensitive genes. To identify targets of HP0165-HP0166, the promoter regions of genes up-regulated by a low pH were grouped based on sequence similarity. Probes for promoter sequences representing each group were subjected to electrophoretic mobility shift assays (EMSA) with recombinant HP0166-His(6) or a mutated response regulator, HP0166-D52N-His(6), that can specifically determine the role of phosphorylation of HP0166 in binding (including a control EMSA with in-vitro-phosphorylated HP0166-His(6)). Nineteen of 45 promoter-regulatory regions were found to interact with HP0166-His(6). Seven promoters for genes encoding alpha-carbonic anhydrase, omp11, fecD, lpp20, hypA, and two with unknown function (pHP1397-1396 and pHP0654-0675) were clustered in gene group A, which may respond to changes in the periplasmic pH at a constant cytoplasmic pH and showed phosphorylation-dependent binding in EMSA with HP0166-D52N-His(6). Twelve promoters were clustered in groups B and C whose up-regulation likely also depends on a reduction of the cytoplasmic pH at a medium pH of 5.5 or 4.5. Most of the target promoters in groups B and C showed phosphorylation-dependent binding with HP0166-D52N-His(6), but promoters for ompR (pHP0166-0162), pHP0682-0681, and pHP1288-1289 showed phosphorylation-independent binding. These findings, combined with DNase I footprinting, suggest that HP0165-0166 is an acid-responsive signaling system affecting the expression of pH-sensitive genes. Regulation of these genes responds either to a decrease in the periplasmic pH alone (HP0165 dependent) or also to a decrease in the cytoplasmic pH (HP0165 independent).
Figures




References
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Baumgartner, H. K., and M. H. Montrose. 2004. Regulated alkali secretion acts in tandem with unstirred layers to regulate mouse gastric surface pH. Gastroenterology 126:774-783. - PubMed
-
- Benoit, S., N. Mehta, G. Wang, M. Gatlin, and R. J. Maier. 2004. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb. Pathog. 36:153-157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

