A caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase
- PMID: 16484194
- PMCID: PMC1426549
- DOI: 10.1128/JB.188.5.1835-1846.2006
A caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase
Abstract
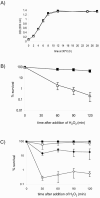
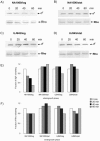
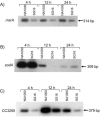
Alternative sigma factors of the extracytoplasmic function (ECF) subfamily are important regulators of stress responses in bacteria and have been implicated in the control of homeostasis of the extracytoplasmic compartment of the cell. This work describes the characterization of sigF, encoding 1 of the 13 members of this subfamily identified in Caulobacter crescentus. A sigF-null strain was obtained and shown to be severely impaired in resistance to oxidative stress, caused by hydrogen peroxide treatment, exclusively during the stationary phase. Although sigF mRNA levels decrease in stationary-phase cells, the amount of sigma(F) protein is greatly increased at this stage, indicating a posttranscriptional control. Data obtained indicate that the FtsH protease is either directly or indirectly involved in the control of sigma(F) levels, as cells lacking this enzyme present larger amounts of the sigma factor. Increased stability of sigma(F) protein in stationary-phase cells of the parental strain and in exponential-phase cells of the ftsH-null strain is also demonstrated. Transcriptome analysis of the sigF-null strain led to the identification of eight genes regulated by sigma(F) during the stationary phase, including sodA and msrA, which are known to be involved in oxidative stress response.
Figures



References
-
- Allen, H. L. 1971. Primary productivity, chemoorganotrophy, and nutritional interactions of epiphytic algae and bacteria on macrophytes in the littoral of a lake. Ecol. Monogr. 41:97-127.
-
- Anderson, D. K., N. Ohta, J. Wu, and A. Newton. 1995. Regulation of the Caulobacter crescentus rpoN gene and function of the purified sigma 54 in flagellar gene transcription. Mol. Gen. Genet. 246:697-706. - PubMed
-
- Blaszczak, A., C. Georgopoulos, and K. Liberek. 1999. On the mechanism of FtsH-dependent degradation of the sigma 32 transcriptional regulator of Escherichia coli and the role of the Dnak chaperone machine. Mol. Microbiol. 31:157-166. - PubMed
-
- Browning, D. F., D. E. Whitworth, and D. A. Hodgson. 2003. Light-induced carotenogenesis in Myxococcus xanthus: functional characterization of the ECF sigma factor CarQ and antisigma factor CarR. Mol. Microbiol. 48:237-251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

