Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings
- PMID: 16484196
- PMCID: PMC1426569
- DOI: 10.1128/JB.188.5.1856-1865.2006
Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings
Abstract
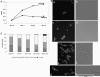
FtsZ, a bacterial homolog of tubulin, forms a structural element called the FtsZ ring (Z ring) at the predivisional midcell site and sets up a scaffold for the assembly of other cell division proteins. The genetic aspects of FtsZ-catalyzed cell division and its assembly dynamics in Mycobacterium tuberculosis are unknown. Here, with an M. tuberculosis strain containing FtsZ(TB) tagged with green fluorescent protein as the sole source of FtsZ, we examined FtsZ structures under various growth conditions. We found that midcell Z rings are present in approximately 11% of actively growing cells, suggesting that the low frequency of Z rings is reflective of their slow growth rate. Next, we showed that SRI-3072, a reported FtsZ(TB) inhibitor, disrupted Z-ring assembly and inhibited cell division and growth of M. tuberculosis. We also showed that M. tuberculosis cells grown in macrophages are filamentous and that only a small fraction had midcell Z rings. The majority of filamentous cells contained nonring, spiral-like FtsZ structures along their entire length. The levels of FtsZ in bacteria grown in macrophages or in broth were comparable, suggesting that Z-ring formation at midcell sites was compromised during intracellular growth. Our results suggest that the intraphagosomal milieu alters the expression of M. tuberculosis genes affecting Z-ring formation and thereby cell division.
Figures


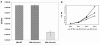



References
-
- Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. - PubMed
-
- Bramhill, D. 1997. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13:395-424. - PubMed
-
- Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

