Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice
- PMID: 16484369
- PMCID: PMC1450143
- DOI: 10.1073/pnas.0511189103
Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice
Abstract
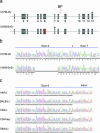
Disrupted-In-Schizophrenia (DISC1) is a leading candidate schizophrenia susceptibility gene. Here, we describe a deletion variant in mDisc1 specific to the 129S6/SvEv strain that introduces a termination codon at exon 7, abolishes production of the full-length protein, and impairs working memory performance when transferred to the C57BL/6J genetic background. Our findings provide insights into how DISC1 variation contributes to schizophrenia susceptibility in humans and the behavioral divergence between 129S6/SvEv and C57BL/6J mouse strains and have implications for modeling psychiatric diseases in mice.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
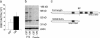

References
-
- Gogos J. A., Gerber D. J. Trends Pharmacol. Sci. 2006 in press. - PubMed
-
- Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., Devon R. S., Clair D. M., Muir W. J., Blackwood D. H., et al. Hum. Mol. Genet. 2000;9:1415–1423. - PubMed
-
- Ekelund J., Hennah W., Hiekkalinna T., Parker A., Meyer J., Lonnqvist J., Peltonen L. Mol. Psychiatry. 2004;9:1037–1041. - PubMed
-
- Hennah W., Tuulio-Henriksson A., Paunio T., Ekelund J., Varilo T., Partonen T., Cannon T. D., Lonnqvist J., Peltonen L. Mol. Psychiatry. 2005;10:1097–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

