Structure modeling of all identified G protein-coupled receptors in the human genome
- PMID: 16485037
- PMCID: PMC1364505
- DOI: 10.1371/journal.pcbi.0020013
Structure modeling of all identified G protein-coupled receptors in the human genome
Erratum in
- PLoS Comput Biol. 2006 Mar;2(3):e29
Abstract
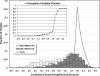
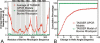
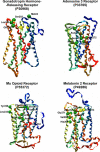
G protein-coupled receptors (GPCRs), encoded by about 5% of human genes, comprise the largest family of integral membrane proteins and act as cell surface receptors responsible for the transduction of endogenous signal into a cellular response. Although tertiary structural information is crucial for function annotation and drug design, there are few experimentally determined GPCR structures. To address this issue, we employ the recently developed threading assembly refinement (TASSER) method to generate structure predictions for all 907 putative GPCRs in the human genome. Unlike traditional homology modeling approaches, TASSER modeling does not require solved homologous template structures; moreover, it often refines the structures closer to native. These features are essential for the comprehensive modeling of all human GPCRs when close homologous templates are absent. Based on a benchmarked confidence score, approximately 820 predicted models should have the correct folds. The majority of GPCR models share the characteristic seven-transmembrane helix topology, but 45 ORFs are predicted to have different structures. This is due to GPCR fragments that are predominantly from extracellular or intracellular domains as well as database annotation errors. Our preliminary validation includes the automated modeling of bovine rhodopsin, the only solved GPCR in the Protein Data Bank. With homologous templates excluded, the final model built by TASSER has a global C(alpha) root-mean-squared deviation from native of 4.6 angstroms, with a root-mean-squared deviation in the transmembrane helix region of 2.1 angstroms. Models of several representative GPCRs are compared with mutagenesis and affinity labeling data, and consistent agreement is demonstrated. Structure clustering of the predicted models shows that GPCRs with similar structures tend to belong to a similar functional class even when their sequences are diverse. These results demonstrate the usefulness and robustness of the in silico models for GPCR functional analysis. All predicted GPCR models are freely available for noncommercial users on our Web site (http://www.bioinformatics.buffalo.edu/GPCR).
Conflict of interest statement
Figures
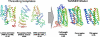

References
-
- Watson S, Arkinstall S. The G protein Linked Receptors Facts Book. New York: Academic Press; 1994. 427. p.
-
- Flower DR. Modelling G-protein-coupled receptors for drug design. Biochim Biophys Acta. 1999;1422:207–234. - PubMed
-
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520:97–101. - PubMed
-
- Collins FS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Drews J. Drug discovery: A historical perspective. Science. 2000;287:1960–1964. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

