Dependency map of proteins in the small ribosomal subunit
- PMID: 16485038
- PMCID: PMC1364506
- DOI: 10.1371/journal.pcbi.0020010
Dependency map of proteins in the small ribosomal subunit
Abstract
The assembly of the ribosome has recently become an interesting target for antibiotics in several bacteria. In this work, we extended an analytical procedure to determine native state fluctuations and contact breaking to investigate the protein stability dependence in the 30S small ribosomal subunit of Thermus thermophilus. We determined the causal influence of the presence and absence of proteins in the 30S complex on the binding free energies of other proteins. The predicted dependencies are in overall agreement with the experimentally determined assembly map for another organism, Escherichia coli. We found that the causal influences result from two distinct mechanisms: one is pure internal energy change, the other originates from the entropy change. We discuss the implications on how to target the ribosomal assembly most effectively by suggesting six proteins as targets for mutations or other hindering of their binding. Our results show that by blocking one out of this set of proteins, the association of other proteins is eventually reduced, thus reducing the translation efficiency even more. We could additionally determine the binding dependency of THX--a peptide not present in the ribosome of E. coli--and suggest its assembly path.
Conflict of interest statement
Figures
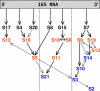
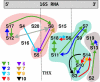

 were marked if the value deviated more than 10% from their most likely value, while for
were marked if the value deviated more than 10% from their most likely value, while for
 this threshold was set to 1%, reflecting the respective order of magnitude of λ1/20.
this threshold was set to 1%, reflecting the respective order of magnitude of λ1/20.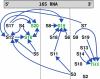

 eigenvector contributions for the two different average interaction values (shown for the worst case of S7).
eigenvector contributions for the two different average interaction values (shown for the worst case of S7).Similar articles
-
All-atom homology model of the Escherichia coli 30S ribosomal subunit.Nat Struct Biol. 2002 Oct;9(10):750-5. doi: 10.1038/nsb841. Nat Struct Biol. 2002. PMID: 12244297
-
Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly.J Bacteriol. 2004 Sep;186(17):5819-25. doi: 10.1128/JB.186.17.5819-5825.2004. J Bacteriol. 2004. PMID: 15317787 Free PMC article.
-
Exploring assembly energetics of the 30S ribosomal subunit using an implicit solvent approach.J Am Chem Soc. 2005 Aug 10;127(31):11125-33. doi: 10.1021/ja052639e. J Am Chem Soc. 2005. PMID: 16076220
-
A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria?RNA Biol. 2004 May;1(1):10-5. Epub 2004 May 19. RNA Biol. 2004. PMID: 17194932 Review.
-
The ribosome through the looking glass.Angew Chem Int Ed Engl. 2003 Aug 4;42(30):3464-86. doi: 10.1002/anie.200200544. Angew Chem Int Ed Engl. 2003. PMID: 12900959 Review.
Cited by
-
Molecular motions as a drug target: mechanistic simulations of anthrax toxin edema factor function led to the discovery of novel allosteric inhibitors.Toxins (Basel). 2012 Aug;4(8):580-604. doi: 10.3390/toxins4080580. Epub 2012 Jul 31. Toxins (Basel). 2012. PMID: 23012649 Free PMC article. Review.
-
Allosteric activation of Bordetella pertussis adenylyl cyclase by calmodulin: molecular dynamics and mutagenesis studies.J Biol Chem. 2014 Jul 25;289(30):21131-41. doi: 10.1074/jbc.M113.530410. J Biol Chem. 2014. PMID: 24907274 Free PMC article.
-
Biophysical studies of bacterial ribosome assembly.Curr Opin Struct Biol. 2008 Jun;18(3):299-304. doi: 10.1016/j.sbi.2008.05.001. Epub 2008 Jun 7. Curr Opin Struct Biol. 2008. PMID: 18541423 Free PMC article. Review.
-
A computational investigation on the connection between dynamics properties of ribosomal proteins and ribosome assembly.PLoS Comput Biol. 2012;8(5):e1002530. doi: 10.1371/journal.pcbi.1002530. Epub 2012 May 24. PLoS Comput Biol. 2012. PMID: 22654657 Free PMC article.
-
Mechanical transduction of cytoplasmic-to-transmembrane-domain movements in a hyperpolarization-activated cyclic nucleotide-gated cation channel.J Biol Chem. 2018 Aug 17;293(33):12908-12918. doi: 10.1074/jbc.RA118.002139. Epub 2018 Jun 23. J Biol Chem. 2018. PMID: 29936413 Free PMC article.
References
-
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. - PubMed
-
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. - PubMed
-
- Mizushima S, Nomura M. Assembly mapping of the 30S ribosomal proteins from E. Coli . Nature. 1970;226:1214–1218. - PubMed
-
- Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of the 30S ribosomal proteins from Escherichia coli: Further studies. J Biol Chem. 1974;249:3103–3111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

