Porcine noroviruses related to human noroviruses
- PMID: 16485473
- PMCID: PMC3367634
- DOI: 10.3201/eid1112.050485
Porcine noroviruses related to human noroviruses
Abstract
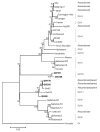
Detection of genogroup II (GII) norovirus (NoV) RNA from adult pigs in Japan and Europe and GII NoV antibodies in US swine raises public health concerns about zoonotic transmission of porcine NoVs to humans, although no NoVs have been detected in US swine. To detect porcine NoVs and to investigate their genetic diversity and relatedness to human NoVs, 275 fecal samples from normal US adult swine were screened by reverse transcription-polymerase chain reaction with calicivirus universal primers. Six samples were positive for NoV. Based on sequence analysis of 3 kb on the 3' end of 5 porcine NoVs, 3 genotypes in GII and a potential recombinant were identified. One genotype of porcine NoVs was genetically and antigenically related to human NoVs and replicated in gnotobiotic pigs. These results raise concerns of whether subclinically infected adult swine may be reservoirs of new human NoVs or if porcine/human GII recombinants could emerge.
Figures




References
-
- Green KY, Chanock RM, Kapikian AZ. Human caliciviruses. In: Knipe DM, Howley PM, editors. Fields virology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 841–74.
-
- Belliot G, Sosnovtsev SV, Mitra T, Hammer C, Garfield M, Green KY. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J Virol. 2003;77:10957–74. 10.1128/JVI.77.20.10957-10974.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
