Intergenogroup recombination in sapoviruses
- PMID: 16485479
- PMCID: PMC3367643
- DOI: 10.3201/eid1112.050722
Intergenogroup recombination in sapoviruses
Abstract
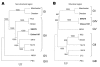
Sapovirus, a member of the family Caliciviridae, is an etiologic agent of gastroenteritis in humans and pigs. Analyses of the complete genome sequences led us to identify the first sapovirus intergenogroup recombinant strain. Phylogenetic analysis of the nonstructural region (i.e., genome start to capsid start) grouped this strain into genogroup II, whereas the structural region (i.e., capsid start to genome end) grouped this strain into genogroup IV. We found that a recombination event occurred at the polymerase and capsid junction. This is the first report of intergenogroup recombination for any calicivirus and highlights a possible route of zoonoses because sapovirus strains that infect pig species belong to genogroup III.
Figures


References
-
- Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–43. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
