Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing
- PMID: 16489123
- PMCID: PMC1383642
- DOI: 10.1105/tpc.105.038240
Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing
Abstract
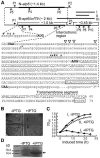
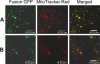
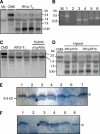
Cytoplasmic male sterility (CMS) and nucleus-controlled fertility restoration are widespread plant reproductive features that provide useful tools to exploit heterosis in crops. However, the molecular mechanism underlying this kind of cytoplasmic-nuclear interaction remains unclear. Here, we show in rice (Oryza sativa) with Boro II cytoplasm that an abnormal mitochondrial open reading frame, orf79, is cotranscribed with a duplicated atp6 (B-atp6) gene and encodes a cytotoxic peptide. Expression of orf79 in CMS lines and transgenic rice plants caused gametophytic male sterility. Immunoblot analysis showed that the ORF79 protein accumulates specifically in microspores. Two fertility restorer genes, Rf1a and Rf1b, were identified at the classical locus Rf-1 as members of a multigene cluster that encode pentatricopeptide repeat proteins. RF1A and RF1B are both targeted to mitochondria and can restore male fertility by blocking ORF79 production via endonucleolytic cleavage (RF1A) or degradation (RF1B) of dicistronic B-atp6/orf79 mRNA. In the presence of both restorers, RF1A was epistatic over RF1B in the mRNA processing. We have also shown that RF1A plays an additional role in promoting the editing of atp6 mRNAs, independent of its cleavage function.
Figures




References
-
- Akagi, H., Nakamura, A., Sawada, R., and Oka, M. (1995). Genetic diagnosis of cytoplasmic male sterile cybrid plants of rice. Theor. Appl. Genet. 90 948–951. - PubMed
-
- Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., and Fujimura, T. (2004). Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108 1449–1457. - PubMed
-
- Akagi, H., Sakamoto, M., Shinjyo, C., Shimada, H., and Fujimura, T. (1994). A unique sequence located downstream from the rice mitochondrial apt6 may cause male sterility. Curr. Genet. 25 52–58. - PubMed
-
- Akagi, H., Yokozeki, Y., Inagaki, A., Nakamura, A., and Fujimura, T. (1996). A codominant DNA marker closely linked to the rice nuclear restorer gene, Rf-1, identified with inter-SSR fingerprinting. Genome 39 1205–1209. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

