Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli
- PMID: 16489229
- PMCID: PMC1456385
- DOI: 10.1534/genetics.105.052373
Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli
Abstract
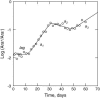
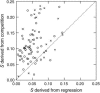
The conventional model of adaptation in asexual populations implies sequential fixation of new beneficial mutations via rare selective sweeps that purge all variation and preserve the clonal genotype. However, in large populations multiple beneficial mutations may co-occur, causing competition among them, a phenomenon called "clonal interference." Clonal interference is thus expected to lead to longer fixation times and larger fitness effects of mutations that ultimately become fixed, as well as to a genetically more diverse population. Here, we study the significance of clonal interference in populations consisting of mixtures of differently marked wild-type and mutator strains of Escherichia coli that adapt to a minimal-glucose environment for 400 generations. We monitored marker frequencies during evolution and measured the competitive fitness of random clones from each marker state after evolution. The results demonstrate the presence of multiple beneficial mutations in these populations and slower and more erratic invasion of mutants than expected by the conventional model, showing the signature of clonal interference. We found that a consequence of clonal interference is that fitness estimates derived from invasion trajectories were less than half the magnitude of direct estimates from competition experiments, thus revealing fundamental problems with this fitness measure. These results force a reevaluation of the conventional model of periodic selection for asexual microbes.
Figures



References
-
- Adams, J., 2004. Microbial evolution in laboratory environments. Res. Microbiol. 155: 311–318. - PubMed
-
- Atwood, K. C., L. K. Schneider and F. J. Ryan, 1951. b Selective mechanisms in bacteria. Cold Spring Harbor Symp. Quant. Biol. 16: 345–355. - PubMed
-
- Chao, L., and E. C. Cox, 1983. Competition between high and low mutating strains of Escherichia coli. Evolution 37: 125–134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

