Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization
- PMID: 16489232
- PMCID: PMC1461447
- DOI: 10.1534/genetics.105.049726
Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization
Abstract
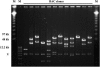
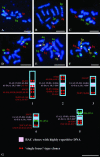
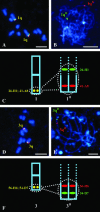
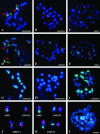
As part of an initiative to develop Brachypodium distachyon as a genomic "bridge" species between rice and the temperate cereals and grasses, a BAC library has been constructed for the two diploid (2n = 2x = 10) genotypes, ABR1 and ABR5. The library consists of 9100 clones, with an approximate average insert size of 88 kb, representing 2.22 genome equivalents. To validate the usefulness of this species for comparative genomics and gene discovery in its larger genome relatives, the library was screened by PCR using primers designed on previously mapped rice and Poaceae sequences. Screening indicated a degree of synteny between these species and B. distachyon, which was confirmed by fluorescent in situ hybridization of the marker-selected BACs (BAC landing) to the 10 chromosome arms of the karyotype, with most of the BACs hybridizing as single loci on known chromosomes. Contiguous BACs colocalized on individual chromosomes, thereby confirming the conservation of genome synteny and proving that B. distachyon has utility as a temperate grass model species alternative to rice.
Figures
References
-
- Allouis, S., G. Moore, A. Bellec, R. Shah, P. Faivre Rampant et al., 2003. Construction and characterisation of a hexaploid wheat (Triticum aestivum L.) BAC library from the reference germplasm ‘Chinese Spring.’ Cereal Res. Commun. 31: 331–338.
-
- Aragon-Alcaide, L., T. Miller, T. Schwarzacher, S. Reader and G. Moore, 1996. A cereal centromeric sequence. Chromosoma 105: 261–268. - PubMed
-
- Armstead, I. P., L. B. Turner, I. P. King, A. J. Cairns and M. O. Humphreys, 2002. Comparison and integration of genetic maps generated from F-2 and BC1-type mapping populations in perennial ryegrass. Plant Breed. 121: 501–507.
-
- Armstead, I. P., J. A. Harper, L. B. Turner, L. Skot, I. P. King et al., 2006. Introgression of crown rust (Puccinia coronata) resistance from meadow fescue (Festuca pratensis) into Italian ryegrass (Lolium multiflorum): genetic mapping and identification of associated molecular markers. Plant Pathol. 55: 62–67. - PubMed
-
- Bablak, P., J. Draper, M. R. Davey and P. T. Lynch, 1995. Plant regeneration and micropropagation of Brachypodium distachyon. Plant Cell Tissue Organ Cult. 42: 97–107.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

