N6-methyl-adenine: an epigenetic signal for DNA-protein interactions
- PMID: 16489347
- PMCID: PMC2755769
- DOI: 10.1038/nrmicro1350
N6-methyl-adenine: an epigenetic signal for DNA-protein interactions
Abstract
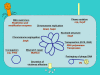
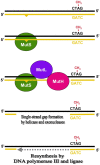
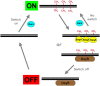
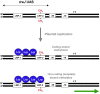
N(6)-methyl-adenine is found in the genomes of bacteria, archaea, protists and fungi. Most bacterial DNA adenine methyltransferases are part of restriction-modification systems. Certain groups of Proteobacteria also harbour solitary DNA adenine methyltransferases that provide signals for DNA-protein interactions. In gamma-proteobacteria, Dam methylation regulates chromosome replication, nucleoid segregation, DNA repair, transposition of insertion elements and transcription of specific genes. In Salmonella, Haemophilus, Yersinia and Vibrio species and in pathogenic Escherichia coli, Dam methylation is required for virulence. In alpha-proteobacteria, CcrM methylation regulates the cell cycle in Caulobacter, Rhizobium and Agrobacterium, and has a role in Brucella abortus infection.
Figures


References
-
- Cheng X. Structure and function of DNA methyltransferases. Annu Rev Biophys Biomol Struct. 1995;24:293–318. - PubMed
-
- Marinus MG. Methylation of DNA. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D. C: 1996. pp. 782–791. A comprehensive summary of the first two decades of research on Dam methylation.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

