Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR
- PMID: 16491076
- PMCID: PMC3001192
- DOI: 10.1038/ni1311
Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR
Abstract
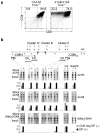
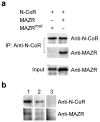
Coreceptor expression is tightly regulated during thymocyte development. Deletion of specific Cd8 enhancers leads to variegated expression of CD8alphabeta heterodimers in double-positive thymocytes. Here we show CD8 variegation is correlated with an epigenetic 'off' state, linking Cd8 enhancer function with chromatin remodeling of the adjacent genes Cd8a and Cd8b1 (Cd8). The zinc finger protein MAZR bound the Cd8 enhancer and interacted with the nuclear receptor corepressor N-CoR complex in double-negative thymocytes. MAZR was downregulated in double-positive and CD8 single-positive thymocytes. 'Enforced' expression of MAZR led to impaired Cd8 activation and variegated CD8 expression. Our results demonstrate epigenetic control of the Cd8 loci and identify MAZR as an important regulator of Cd8 expression.
Figures






References
-
- Fisher AG. Cellular identity and lineage choice. Nat Rev Immunol. 2002;2:977–982. - PubMed
-
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. - PubMed
-
- Smale ST. The establishment and maintenance of lymphocyte identity through gene silencing. Nat Immunol. 2003;4:607–615. - PubMed
-
- Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

