Insertion and assembly of membrane proteins via simulation
- PMID: 16492056
- PMCID: PMC4618310
- DOI: 10.1021/ja0569104
Insertion and assembly of membrane proteins via simulation
Abstract
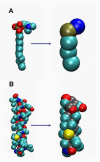
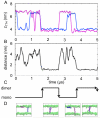
Interactions of lipids are central to the folding and stability of membrane proteins. Coarse-grained molecular dynamics simulations have been used to reveal the mechanisms of self-assembly of protein/membrane and protein/detergent complexes for representatives of two classes of membrane protein, namely, glycophorin (a simple alpha-helical bundle) and OmpA (a beta-barrel). The accuracy of the coarse-grained simulations is established via comparison with the equivalent atomistic simulations of self-assembly of protein/detergent micelles. The simulation of OmpA/bilayer self-assembly reveals how a folded outer membrane protein can be inserted in a bilayer. The glycophorin/bilayer simulation supports the two-state model of membrane folding, in which transmembrane helix insertion precedes dimer self-assembly within a bilayer. The simulations also suggest that a dynamic equilibrium exists between the glycophorin helix monomer and dimer within a bilayer. The simulated glycophorin helix dimer is remarkably close in structure to that revealed by NMR. Thus, coarse-grained methods may help to define mechanisms of membrane protein (re)folding and will prove suitable for simulation of larger scale dynamic rearrangements of biological membranes.
Figures





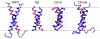

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

