Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen
- PMID: 16492767
- PMCID: PMC1413915
- DOI: 10.1073/pnas.0511224103
Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen
Abstract
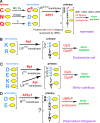
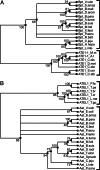
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. Primary destabilizing N-terminal residues (Nd(p)) are recognized directly by the targeting machinery. The recognition of secondary destabilizing N-terminal residues (Nd(s)) is preceded by conjugation of an Nd(p) residue to Nd(s) of a polypeptide substrate. In eukaryotes, ATE1-encoded arginyl-transferases (R(D,E,C*)-transferases) conjugate Arg (R), an Nd(p) residue, to Nd(s) residues Asp (D), Glu (E), or oxidized Cys residue (C*). Ubiquitin ligases recognize the N-terminal Arg of a substrate and target the (ubiquitylated) substrate to the proteasome. In prokaryotes such as Escherichia coli, Nd(p) residues Leu (L) or Phe (F) are conjugated, by the aat-encoded Leu/Phe-transferase (L/F(K,R)-transferase), to N-terminal Arg or Lys, which are Nd(s) in prokaryotes but Nd(p) in eukaryotes. In prokaryotes, substrates bearing the Nd(p) residues Leu, Phe, Trp, or Tyr are degraded by the proteasome-like ClpAP protease. Despite enzymological similarities between eukaryotic R(D,E,C*)-transferases and prokaryotic L/F(K,R)-transferases, there is no significant sequelogy (sequence similarity) between them. We identified an aminoacyl-transferase, termed Bpt, in the human pathogen Vibrio vulnificus. Although it is a sequelog of eukaryotic R(D,E,C*)-transferases, this prokaryotic transferase exhibits a "hybrid" specificity, conjugating Nd(p) Leu to Nd(s) Asp or Glu. Another aminoacyl-transferase, termed ATEL1, of the eukaryotic pathogen Plasmodium falciparum, is a sequelog of prokaryotic L/F(K,R)-transferases (Aat), but has the specificity of eukaryotic R(D,E,C*)-transferases (ATE1). Phylogenetic analysis suggests that the substrate specificity of R-transferases arose by two distinct routes during the evolution of eukaryotes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Varshavsky A. Trends Biochem. Sci. 2005;30:283–286. - PubMed
-
- Hershko A., Ciechanover A., Varshavsky A. Nat. Med. 2000;10:1073–1081. - PubMed
-
- Pickart C. Cell. 2004;116:181–190. - PubMed
-
- Petroski M. D., Deshaies R. J. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. - PubMed
-
- Baumeister W., Walz J., Zühl F., Seemüller E. Cell. 1998;92:367–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

