Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation
- PMID: 16493008
- PMCID: PMC2875852
- DOI: 10.1182/blood-2005-09-3596
Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation
Abstract
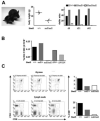
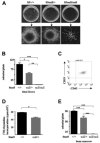
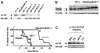
The Stat5 transcription factors Stat5a and Stat5b have been implicated in lymphoid development and transformation. Most studies have employed Stat5a/b-deficient mice where gene targeting disrupted the first protein-coding exon, resulting in the expression of N-terminally truncated forms of Stat5a/b (Stat5a/b(DeltaN/DeltaN) mice). We have now reanalyzed lymphoid development in Stat5a/b(null/null) mice having a complete deletion of the Stat5a/b gene locus. The few surviving Stat5a/b(null/null) mice lacked CD8(+) T lymphocytes. A massive reduction of CD8(+) T cells was also found in Stat5a/b(fl/fl) lck-cre transgenic animals. While gammadelta T-cell receptor-positive (gammadeltaTCR(+)) cells were expressed at normal levels in Stat5a/b(DeltaN/DeltaN) mice, they were completely absent in Stat5a/b(null/null) animals. Moreover, B-cell maturation was abrogated at the pre-pro-B-cell stage in Stat5a/b(null/null) mice, whereas Stat5a/b(DeltaN/DeltaN) B-lymphoid cells developed to the early pro-B-cell stage. In vitro assays using fetal liver-cell cultures confirmed this observation. Most strikingly, Stat5a/b(null/null) cells were resistant to transformation and leukemia development induced by Abelson oncogenes, whereas Stat5a/b(DeltaN/DeltaN)-derived cells readily transformed. These findings show distinct lymphoid defects for Stat5a/b(DeltaN/DeltaN) and Stat5a/b(null/null) mice and define a novel functional role for the N-termini of Stat5a/b in B-lymphoid transformation.
Figures



References
-
- Calo V, Migliavacca M, Bazan V, et al. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. - PubMed
-
- Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. - PubMed
-
- Murray R. Physiologic roles of interleukin-2, interleukin-4, and interleukin-7. Curr Opin Hematol. 1996;3:230–234. - PubMed
-
- Moriggl R, Topham DJ, Teglund S, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. - PubMed
-
- Gouilleux-Gruart V, Debierre-Grockiego F, Gouilleux F, et al. Activated Stat related transcription factors in acute leukemia. Leuk Lymphoma. 1997;28:83–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

