A map of recent positive selection in the human genome
- PMID: 16494531
- PMCID: PMC1382018
- DOI: 10.1371/journal.pbio.0040072
A map of recent positive selection in the human genome
Erratum in
- PLoS Biol. 2006 Apr;4(4):e154
- PLoS Biol. 2007 Jun;5(6):e147
Abstract
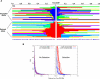
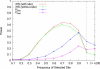
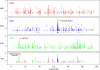
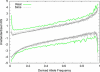
The identification of signals of very recent positive selection provides information about the adaptation of modern humans to local conditions. We report here on a genome-wide scan for signals of very recent positive selection in favor of variants that have not yet reached fixation. We describe a new analytical method for scanning single nucleotide polymorphism (SNP) data for signals of recent selection, and apply this to data from the International HapMap Project. In all three continental groups we find widespread signals of recent positive selection. Most signals are region-specific, though a significant excess are shared across groups. Contrary to some earlier low resolution studies that suggested a paucity of recent selection in sub-Saharan Africans, we find that by some measures our strongest signals of selection are from the Yoruba population. Finally, since these signals indicate the existence of genetic variants that have substantially different fitnesses, they must indicate loci that are the source of significant phenotypic variation. Though the relevant phenotypes are generally not known, such loci should be of particular interest in mapping studies of complex traits. For this purpose we have developed a set of SNPs that can be used to tag the strongest approximately 250 signals of recent selection in each population.
Figures
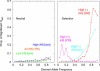
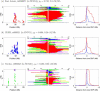

Comment in
-
Being positive about selection.PLoS Biol. 2006 Mar;4(3):e87. doi: 10.1371/journal.pbio.0040087. Epub 2006 Mar 14. PLoS Biol. 2006. PMID: 16524343 Free PMC article.
-
Clues to our past: mining the human genome for signs of recent selection.PLoS Biol. 2006 Mar;4(3):e94. doi: 10.1371/journal.pbio.0040094. Epub 2006 Mar 7. PLoS Biol. 2006. PMID: 20076547 Free PMC article. No abstract available.
References
-
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. - PubMed
-
- Jobling MA, Hurles ME, Tyler-Smith C. Human evolutionary genetics: Origins, peoples and disease. New York: Garland Science; 2004. 523 pp.
-
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, et al. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. - PubMed
-
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

