Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease
- PMID: 16495249
- PMCID: PMC1426435
- DOI: 10.1128/AAC.50.3.899-909.2006
Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease
Abstract
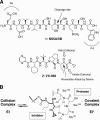
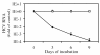
VX-950 is a potent, selective, peptidomimetic inhibitor of the hepatitis C virus (HCV) NS3-4A serine protease, and it demonstrated excellent antiviral activity both in genotype 1b HCV replicon cells (50% inhibitory concentration [IC50] = 354 nM) and in human fetal hepatocytes infected with genotype 1a HCV-positive patient sera (IC50 = 280 nM). VX-950 forms a covalent but reversible complex with the genotype 1a HCV NS3-4A protease in a slow-on, slow-off process with a steady-state inhibition constant (K(i)*) of 7 nM. Dissociation of the covalent enzyme-inhibitor complex of VX-950 and genotype 1a HCV protease has a half-life of almost an hour. A >4-log10 reduction in the HCV RNA levels was observed after a 2-week incubation of replicon cells with VX-950, with no rebound of viral RNA observed after withdrawal of the inhibitor. In several animal species, VX-950 exhibits a favorable pharmacokinetic profile with high exposure in the liver. In a recently developed HCV protease mouse model, VX-950 showed excellent inhibition of HCV NS3-4A protease activity in the liver. Therefore, the overall preclinical profile of VX-950 supports its candidacy as a novel oral therapy against hepatitis C.
Figures


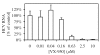

References
-
- De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. - PubMed
-
- Di Marco, S., M. Rizzi, C. Volpari, M. A. Walsh, F. Narjes, S. Colarusso, R. De Francesco, V. G. Matassa, and M. Sollazzo. 2000. Inhibition of the hepatitis C virus NS3/4A protease. The crystal structures of two protease-inhibitor complexes. J. Biol. Chem. 275:7152-7157. - PubMed
-
- Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

